bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: Microviridae_proteins.fasta
575 sequences; 147,441 total letters
Query= Contig-24_CDS_annotation_glimmer3.pl_2_3
Length=591
Score E
Sequences producing significant alignments: (Bits) Value
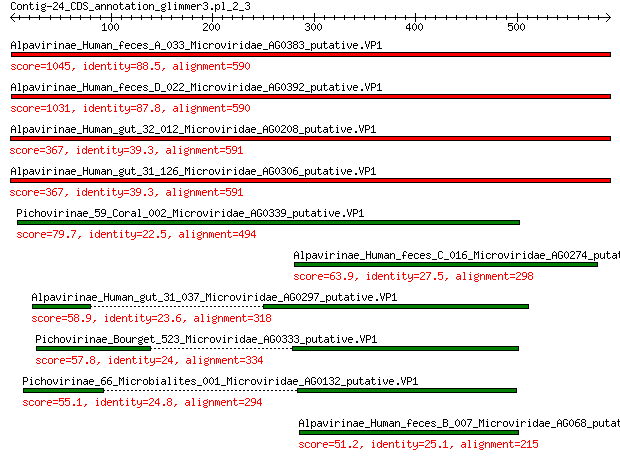
Alpavirinae_Human_feces_A_033_Microviridae_AG0383_putative.VP1 1045 0.0
Alpavirinae_Human_feces_D_022_Microviridae_AG0392_putative.VP1 1031 0.0
Alpavirinae_Human_gut_32_012_Microviridae_AG0208_putative.VP1 367 3e-120
Alpavirinae_Human_gut_31_126_Microviridae_AG0306_putative.VP1 367 3e-120
Pichovirinae_59_Coral_002_Microviridae_AG0339_putative.VP1 79.7 1e-17
Alpavirinae_Human_feces_C_016_Microviridae_AG0274_putative.VP1 63.9 2e-12
Alpavirinae_Human_gut_31_037_Microviridae_AG0297_putative.VP1 58.9 7e-11
Pichovirinae_Bourget_523_Microviridae_AG0333_putative.VP1 57.8 1e-10
Pichovirinae_66_Microbialites_001_Microviridae_AG0132_putative.VP1 55.1 1e-09
Alpavirinae_Human_feces_B_007_Microviridae_AG068_putative.VP1 51.2 1e-08
> Alpavirinae_Human_feces_A_033_Microviridae_AG0383_putative.VP1
Length=590
Score = 1045 bits (2701), Expect = 0.0, Method: Compositional matrix adjust.
Identities = 522/590 (88%), Positives = 553/590 (94%), Gaps = 0/590 (0%)
Query 2 MFLSRKRNKKSRFKLFSGNPTSASWGTLIPTNVTRVVAGDDFSFQPGVGVQALPIVAPFM 61
MFLSRKRNKKSRFKLFSGNPTSASWGTLIPTNVTRVVAGDDFSFQPGVGVQALPIVAPFM
Sbjct 1 MFLSRKRNKKSRFKLFSGNPTSASWGTLIPTNVTRVVAGDDFSFQPGVGVQALPIVAPFM 60
Query 62 GSVCVKKEYFFIPDRIYNVDRQLNFQGVTDTPNTVYKPSIAPPIPFDISKPSGDAISFPV 121
GSVCVKKEYFFIPDRIYN++RQLNFQGVTDTPNTVYKPS+APPIPFDIS SG+ +S PV
Sbjct 61 GSVCVKKEYFFIPDRIYNIERQLNFQGVTDTPNTVYKPSMAPPIPFDISVASGEEVSIPV 120
Query 122 SALQTSVPSGSLGAIVGPGSLADYMGEAPGSIVTGVIDLTPYVGyidiyynyylnqqydl 181
S +QT +P SLG IVGPGSLADYMGEAPGSIVTGVIDLTPY+GYIDIYYNYY+NQQYDL
Sbjct 121 SDVQTELPLVSLGDIVGPGSLADYMGEAPGSIVTGVIDLTPYIGYIDIYYNYYVNQQYDL 180
Query 182 VPTALAGTVSDSAMEYPYYLSVFELETYLRNIKTKSNFSPAIREGNSVSYSTNVKAALNA 241
VPT+LAGT SDSA+EYPYYLSV ELETYLR IKT N SPAIR +SVSYSTNV+ AL A
Sbjct 181 VPTSLAGTKSDSAVEYPYYLSVSELETYLRTIKTTPNTSPAIRVDDSVSYSTNVEKALQA 240
Query 242 VDSEAFSWYFFAGRQSLFQRGFPSYYLEAWLKTSSFTDAAVDVSTSGNSVSMRNITFASR 301
V S AFSW FF GRQSLFQRGFPSYYLEAWLKTSSF DAAVDVS SG+SVSMRNITFASR
Sbjct 241 VGSYAFSWNFFTGRQSLFQRGFPSYYLEAWLKTSSFVDAAVDVSASGSSVSMRNITFASR 300
Query 302 MQRYMDLAFAGGGRNSDFYESQFDVKLNQDNTCPAFLGSDSFDMNANTLYQTTGFEDSSS 361
MQRYMDLAFAGGGRNSDFYESQFDVKL+QDNTCPAFLGSDSFDMN NTLYQTTGFED+SS
Sbjct 301 MQRYMDLAFAGGGRNSDFYESQFDVKLDQDNTCPAFLGSDSFDMNVNTLYQTTGFEDNSS 360
Query 362 PLGAFSGQLSGGTRFRRRNYHFNDDGYFMEITSIVPRTYYPSYINPTSRQISLGQQYAPA 421
PLGAFSGQLSGGTRFRRRNYHFNDDGYFMEITSIVPR YYPSYINPTSRQISLGQQYAPA
Sbjct 361 PLGAFSGQLSGGTRFRRRNYHFNDDGYFMEITSIVPRVYYPSYINPTSRQISLGQQYAPA 420
Query 422 LDNIAMQGLKASTVFGEVQSLGATSPTYSNSVFAVPGFKLQDTNYVGYEPAWSELMTAVS 481
LDNIAMQGLKASTVFGEVQSLGAT+P+Y+ + +PGFKLQD NY+GYEPAWSELMTAVS
Sbjct 421 LDNIAMQGLKASTVFGEVQSLGATNPSYAENRLIIPGFKLQDFNYIGYEPAWSELMTAVS 480
Query 482 KPHGRLCNDLDYWVLSRDYGRNLASVMDTPAYKSFVSAAGAHVEELALQRLTAFFKRIYV 541
KPHGRLCNDLDYWVLSRDYGRN+A VMD+ AYK F++AAG+ V+EL+LQRLTAFFKRIY+
Sbjct 481 KPHGRLCNDLDYWVLSRDYGRNIAHVMDSKAYKDFIAAAGSGVDELSLQRLTAFFKRIYI 540
Query 542 SPSSCPYILCGDFNYVFYDQRATAENFVLDNVADIVVFREKSKVNVATTL 591
SPSSCPYILCGDFNYVFYDQR TAENF+LDNVADIVVFREKSKVNVATTL
Sbjct 541 SPSSCPYILCGDFNYVFYDQRPTAENFILDNVADIVVFREKSKVNVATTL 590
> Alpavirinae_Human_feces_D_022_Microviridae_AG0392_putative.VP1
Length=589
Score = 1031 bits (2667), Expect = 0.0, Method: Compositional matrix adjust.
Identities = 518/590 (88%), Positives = 548/590 (93%), Gaps = 1/590 (0%)
Query 2 MFLSRKRNKKSRFKLFSGNPTSASWGTLIPTNVTRVVAGDDFSFQPGVGVQALPIVAPFM 61
MFLSRKRNKKSRFKLFSGNPTSASWGTLIPTNVTRV+AGDDFSFQPGVGVQALPIVAPFM
Sbjct 1 MFLSRKRNKKSRFKLFSGNPTSASWGTLIPTNVTRVIAGDDFSFQPGVGVQALPIVAPFM 60
Query 62 GSVCVKKEYFFIPDRIYNVDRQLNFQGVTDTPNTVYKPSIAPPIPFDISKPSGDAISFPV 121
G+VCVKKEYFFIPDRIYN+DRQLNFQGVTDTPNTVYKPS+APPIPFDIS PSG + V
Sbjct 61 GNVCVKKEYFFIPDRIYNIDRQLNFQGVTDTPNTVYKPSMAPPIPFDISAPSG-KVDISV 119
Query 122 SALQTSVPSGSLGAIVGPGSLADYMGEAPGSIVTGVIDLTPYVGyidiyynyylnqqydl 181
S +QT +P SLG IVGPGSLADYMGEAPGSIVTGVIDLTPY+GYIDIYYNYYLNQQ+ L
Sbjct 120 SDVQTDLPLRSLGYIVGPGSLADYMGEAPGSIVTGVIDLTPYIGYIDIYYNYYLNQQFGL 179
Query 182 VPTALAGTVSDSAMEYPYYLSVFELETYLRNIKTKSNFSPAIREGNSVSYSTNVKAALNA 241
VPT+LAGTVSDSAMEYPY+L V ELE YLR IKT N SPAIRE +S SYSTNV AAL A
Sbjct 180 VPTSLAGTVSDSAMEYPYFLDVSELEGYLRAIKTTPNTSPAIREDDSASYSTNVNAALRA 239
Query 242 VDSEAFSWYFFAGRQSLFQRGFPSYYLEAWLKTSSFTDAAVDVSTSGNSVSMRNITFASR 301
V S AF W FF GRQSLFQRGFPSYYLEAWLKTSSF+DAAVD+STSG+SVSMRNITFASR
Sbjct 240 VASNAFLWDFFTGRQSLFQRGFPSYYLEAWLKTSSFSDAAVDISTSGSSVSMRNITFASR 299
Query 302 MQRYMDLAFAGGGRNSDFYESQFDVKLNQDNTCPAFLGSDSFDMNANTLYQTTGFEDSSS 361
MQRYMDLAFAGGGRNSDFYE+QFDVKLNQDNTCPAFLGSDSFDMN NTLYQTTGFED+SS
Sbjct 300 MQRYMDLAFAGGGRNSDFYEAQFDVKLNQDNTCPAFLGSDSFDMNVNTLYQTTGFEDNSS 359
Query 362 PLGAFSGQLSGGTRFRRRNYHFNDDGYFMEITSIVPRTYYPSYINPTSRQISLGQQYAPA 421
PLGAFSGQLSGGTRFRRRNYHFNDDGYFMEITSIVPR YYPSYINPTSRQISLGQQYAPA
Sbjct 360 PLGAFSGQLSGGTRFRRRNYHFNDDGYFMEITSIVPRVYYPSYINPTSRQISLGQQYAPA 419
Query 422 LDNIAMQGLKASTVFGEVQSLGATSPTYSNSVFAVPGFKLQDTNYVGYEPAWSELMTAVS 481
LDNIAMQGLKASTVFGEVQ+L A S TY+N + ++PGFK+QD+NYVGYEPAWSELMTAVS
Sbjct 420 LDNIAMQGLKASTVFGEVQNLSAISTTYANFILSIPGFKVQDSNYVGYEPAWSELMTAVS 479
Query 482 KPHGRLCNDLDYWVLSRDYGRNLASVMDTPAYKSFVSAAGAHVEELALQRLTAFFKRIYV 541
KPHGRLCNDLDYWVLSRDYGRNL+ VMD+ AY FVSAAG +++EL+LQRLTAFFKRIY
Sbjct 480 KPHGRLCNDLDYWVLSRDYGRNLSYVMDSKAYGDFVSAAGTNIDELSLQRLTAFFKRIYE 539
Query 542 SPSSCPYILCGDFNYVFYDQRATAENFVLDNVADIVVFREKSKVNVATTL 591
SPSSCPYILCGDFNYVFYDQR TAENFVLDNVADIVVFREKSKVNVATTL
Sbjct 540 SPSSCPYILCGDFNYVFYDQRPTAENFVLDNVADIVVFREKSKVNVATTL 589
> Alpavirinae_Human_gut_32_012_Microviridae_AG0208_putative.VP1
Length=604
Score = 367 bits (943), Expect = 3e-120, Method: Compositional matrix adjust.
Identities = 232/625 (37%), Positives = 330/625 (53%), Gaps = 57/625 (9%)
Query 1 MMFLSRKRNKKSRFKLFSGNPTSASWGTLIPTNVTRVVAGDDFSFQPGVGVQALPIVAPF 60
MMFL+R RNKKS L +PT+ + G L+P + TR+ AGDD F+P VQA+P+ AP
Sbjct 3 MMFLTRSRNKKSNVNLSHVSPTTIAPGNLVPISFTRIFAGDDLRFEPSAFVQAMPMNAPL 62
Query 61 MGSVCVKKEYFFIPDRIYNVDRQLNFQGVTDTPNTVYKPSIAPPIPFDISKPSGDAISFP 120
+ + EYFF+PDR+YN + ++ GVTD P++V P I P + + D I F
Sbjct 63 VNGFKLCLEYFFVPDRLYNWNLLMDNTGVTDNPDSVKFPQIHAPAEY-----TSDTIKFS 117
Query 121 VSALQTSVPSGS--LGAIVGPGSLADYMGEAPGSIVT-------------------GVID 159
+++ + + + +IV PGSLADY G G T GV+D
Sbjct 118 LNSESAAKGNAARLANSIVQPGSLADYCGFPVGLFPTYEVVLDTDDRNQFCALKLLGVLD 177
Query 160 LTPYVGyidiyynyylnqqydlVPTALAGTVSDSAMEYPYYLSVFELETYLRNIKTKSNF 219
+ + + PT G+ + ++Y SV L ++L +K ++
Sbjct 178 IFYH---YYVNQQIDRFPTASFTPTLNHGSDEERNVDY----SVTMLRSFLDFVKRSADP 230
Query 220 SPAIREGNSVSYSTNVKAALNAVDSEAF-SWYFFAGRQSLFQRGFPSYYLEAWLKTSSFT 278
+ AI + A N + F +W +F R S+FQR P YYLE+WL TS +
Sbjct 231 AGAIGQW----------ATSNPNGNPIFGTWSWFCSRASIFQRCLPPYYLESWLATSGYE 280
Query 279 DA--AVDVSTSGNSVSMRNITFASRMQRYMDLAFAGGGRNSDFYESQFDVKLNQDNTCPA 336
D+ VD+ G S+S RNI S +QR++DLA AGG R SD+ SQFDV + T P
Sbjct 281 DSEIKVDLGADGKSISFRNIAAQSHIQRWLDLALAGGSRYSDYINSQFDVSRLKHTTSPL 340
Query 337 FLGSDSFDMNANTLYQTTGFEDSSSPLGAFSGQLSGGTRFRRRNYHFNDDGYFMEITSIV 396
+LGSD + +N +YQTTG DS+SPLGAF+GQ SGG FRRR+YHF ++GYF+ + S+V
Sbjct 341 YLGSDRQYLGSNVIYQTTGAGDSASPLGAFAGQSSGGETFRRRSYHFGENGYFVVMASLV 400
Query 397 PRTYYPSYINPTSRQISLGQQYAPALDNIAMQGLKASTVFGEVQSLGATSPTYSNSVFAV 456
P Y ++P +R+ +LG Y PALDNIAM+ L V + SL + S ++
Sbjct 401 PDVVYSRGMDPFNREKTLGDVYVPALDNIAMEPLMIEQV-DAIPSLRSLQKVDSVYTLSI 459
Query 457 PGFKLQDTNYVGYEPAWSELMTAVSKPHGRLCNDLDYWVLSRDYGRNLASVMDT-----P 511
KL + +GY PAWS++M VS+ HGRL DL YW+LSRDYG ++ + +T
Sbjct 460 RPDKLIKNSALGYVPAWSKVMQNVSRAHGRLTTDLKYWLLSRDYGVDVDKIKNTGVIGAD 519
Query 512 AYKSFVSA-----AGAHVEELALQRLTAFFKRIYVSPSSCPYILCGDFNYVFYDQRATAE 566
K+ S ++ + L F RI P PY+L +N VF D A+
Sbjct 520 VLKNLASELNQAYQAGYISFEQMDALVTLFSRIQALPEYTPYVLPNAYNDVFADTSNQAQ 579
Query 567 NFVLDNVADIVVFREKSKVNVATTL 591
NFVL + REK KVN TT+
Sbjct 580 NFVLTCTFSMTCNREKGKVNTPTTI 604
> Alpavirinae_Human_gut_31_126_Microviridae_AG0306_putative.VP1
Length=604
Score = 367 bits (943), Expect = 3e-120, Method: Compositional matrix adjust.
Identities = 232/625 (37%), Positives = 330/625 (53%), Gaps = 57/625 (9%)
Query 1 MMFLSRKRNKKSRFKLFSGNPTSASWGTLIPTNVTRVVAGDDFSFQPGVGVQALPIVAPF 60
MMFL+R RNKKS L +PT+ + G L+P + TR+ AGDD F+P VQA+P+ AP
Sbjct 3 MMFLTRSRNKKSNVNLSHVSPTTIAPGNLVPISFTRIFAGDDLRFEPSAFVQAMPMNAPL 62
Query 61 MGSVCVKKEYFFIPDRIYNVDRQLNFQGVTDTPNTVYKPSIAPPIPFDISKPSGDAISFP 120
+ + EYFF+PDR+YN + ++ GVTD P++V P I P + + D I F
Sbjct 63 VNGFKLCLEYFFVPDRLYNWNLLMDNTGVTDNPDSVKFPQIHAPAEY-----TSDTIKFS 117
Query 121 VSALQTSVPSGS--LGAIVGPGSLADYMGEAPGSIVT-------------------GVID 159
+++ + + + +IV PGSLADY G G T GV+D
Sbjct 118 LNSESAAKGNAARLANSIVQPGSLADYCGFPVGLFPTYEVVLDTDDRNQFCALKLLGVLD 177
Query 160 LTPYVGyidiyynyylnqqydlVPTALAGTVSDSAMEYPYYLSVFELETYLRNIKTKSNF 219
+ + + PT G+ + ++Y SV L ++L +K ++
Sbjct 178 IFYH---YYVNQQIDRFPTASFTPTLNHGSDEERNVDY----SVTMLRSFLDFVKRSADP 230
Query 220 SPAIREGNSVSYSTNVKAALNAVDSEAF-SWYFFAGRQSLFQRGFPSYYLEAWLKTSSFT 278
+ AI + A N + F +W +F R S+FQR P YYLE+WL TS +
Sbjct 231 AGAIGQW----------ATSNPNGNPIFGTWSWFCSRASIFQRCLPPYYLESWLATSGYE 280
Query 279 DA--AVDVSTSGNSVSMRNITFASRMQRYMDLAFAGGGRNSDFYESQFDVKLNQDNTCPA 336
D+ VD+ G S+S RNI S +QR++DLA AGG R SD+ SQFDV + T P
Sbjct 281 DSEIKVDLGADGKSISFRNIAAQSHIQRWLDLALAGGSRYSDYINSQFDVSRLKHTTSPL 340
Query 337 FLGSDSFDMNANTLYQTTGFEDSSSPLGAFSGQLSGGTRFRRRNYHFNDDGYFMEITSIV 396
+LGSD + +N +YQTTG DS+SPLGAF+GQ SGG FRRR+YHF ++GYF+ + S+V
Sbjct 341 YLGSDRQYLGSNVIYQTTGAGDSASPLGAFAGQSSGGETFRRRSYHFGENGYFVVMASLV 400
Query 397 PRTYYPSYINPTSRQISLGQQYAPALDNIAMQGLKASTVFGEVQSLGATSPTYSNSVFAV 456
P Y ++P +R+ +LG Y PALDNIAM+ L V + SL + S ++
Sbjct 401 PDVVYSRGMDPFNREKTLGDVYVPALDNIAMEPLMIEQV-DAIPSLRSLQKVDSVYTLSI 459
Query 457 PGFKLQDTNYVGYEPAWSELMTAVSKPHGRLCNDLDYWVLSRDYGRNLASVMDT-----P 511
KL + +GY PAWS++M VS+ HGRL DL YW+LSRDYG ++ + +T
Sbjct 460 RPDKLIKNSALGYVPAWSKVMQNVSRAHGRLTTDLKYWLLSRDYGVDVDKIKNTGVIGAD 519
Query 512 AYKSFVSA-----AGAHVEELALQRLTAFFKRIYVSPSSCPYILCGDFNYVFYDQRATAE 566
K+ S ++ + L F RI P PY+L +N VF D A+
Sbjct 520 VLKNLASELNQAYQAGYISFEQMDALVTLFSRIQALPEYTPYVLPNAYNDVFADTSNQAQ 579
Query 567 NFVLDNVADIVVFREKSKVNVATTL 591
NFVL + REK KVN TT+
Sbjct 580 NFVLTCTFSMTCNREKGKVNTPTTI 604
> Pichovirinae_59_Coral_002_Microviridae_AG0339_putative.VP1
Length=522
Score = 79.7 bits (195), Expect = 1e-17, Method: Compositional matrix adjust.
Identities = 111/512 (22%), Positives = 185/512 (36%), Gaps = 70/512 (14%)
Query 8 RNKKSRFKLFSGNPTSASWGTLIPTNVTRVVAGDDFSFQPGVGVQALPIVAPFMGSVCVK 67
R + + F L S G L+P V V GD F+ + + P++ P M V
Sbjct 11 RPQTNTFDLSHDRKFSGKIGELMPITVMEAVPGDKFNIKATNLTRFAPLITPIMHQASVY 70
Query 68 KEYFFIPDRIYNVDRQLNFQGVTDTPNTVYKPSIAPPIPFDISKPSGDAISFPVSALQTS 127
+FF+P+RI + + G D +A P +FP + T
Sbjct 71 CHFFFVPNRILWSNWEDFISGGED--------GLADP-------------TFPTINITTP 109
Query 128 VPSGSLGAIVGPGSLADYMGEAPGSIVTGVIDLTPYVGyidiyynyylnqqydlVPTALA 187
++GA LADY+G G+ + GV L Y D V L+
Sbjct 110 TNGYAVGA------LADYLGLPTGTQIDGVSALPFAAYQKIYDDYYRDENLIDKVDITLS 163
Query 188 ------------GTVSDSAMEYPYYLSVFELETYLRNIKTKSNFSPAIREGNSVSYSTNV 235
GT+ A ++ Y+ S S I N+ + +T V
Sbjct 164 DGTQSGADAIELGTMRKRAWQHDYFTSALPWTQRGPEATIPLGTSAPITWSNNTATATIV 223
Query 236 KAALNAVDSEAFSWYFFAGRQSLFQRGFPSYYLEAWLKTS-----SFTDAAVDVSTSGNS 290
+ N + Y F G +L+ G S L A L T +D +
Sbjct 224 R---NNTSGNPIAGYTFDGASALYTGG--SGQLIADLPTPVSLDFDNSDHLFADLAGATA 278
Query 291 VSMRNITFASRMQRYMDLAFAGGGRNSDFYESQFDVKLNQDNTC-PAFLGSDSFDMNANT 349
S+ ++ A R+Q +++ GG R + + F V+ + P FLG + + +
Sbjct 279 SSINDLRRAFRLQEWLERNARGGARYIEIIMAHFGVRSSDARLQRPEFLGGSATPITISE 338
Query 350 LYQTTGFEDSSSPLGAFSGQLSGGTRFRRRNYHFNDDGYFMEITSIVPRTYYPSYINPTS 409
+ QT+ +P G +G +Y + GY + I S++P+T Y I
Sbjct 339 VLQTSANNTEPTPQGNMAGHGVSVGSSNYVSYKCEEHGYIIGIMSVMPKTAYQQGIPKHF 398
Query 410 RQISLGQQYAPALDNIAMQGLKASTVFGEVQSLGATSPTYSNSVFAVPGFKLQDTNYVGY 469
+++ Y P+ NI Q + ++ + + DT GY
Sbjct 399 KKLDKFDYYWPSFANIGEQPILNEELYHQNNA--------------------TDTETFGY 438
Query 470 EPAWSELMTAVSKPHGRLCNDLDYWVLSRDYG 501
P ++E S HG + LD+W + R +G
Sbjct 439 TPRYAEYKYIPSTVHGEFRDSLDFWHMGRIFG 470
> Alpavirinae_Human_feces_C_016_Microviridae_AG0274_putative.VP1
Length=641
Score = 63.9 bits (154), Expect = 2e-12, Method: Compositional matrix adjust.
Identities = 82/318 (26%), Positives = 128/318 (40%), Gaps = 54/318 (17%)
Query 281 AVDVST-SGNSVSMRNITFASRMQRYMDLAFAGGGRNSDFYESQFDVKLNQDNTC-PAFL 338
A+D +T +G S+++ + +++Q +MD F GGR D + + + K + P FL
Sbjct 344 ALDSNTDTGFSIAVPELRLKTKIQNWMDRLFISGGRVGDVFRTLWGTKSSAPYVNKPDFL 403
Query 339 GSDSFDMNANTLYQTTGFEDSSSPLGAFSGQLSGG-------TRFRRRNYHFNDDGYFME 391
G +N + + S+S A GQL+ + +Y+ + G FM
Sbjct 404 GVWQASINPSNVRAMA--NGSASGEDANLGQLAACVDRYCDFSDHSGIDYYAKEPGTFML 461
Query 392 ITSIVPRTYYPSYINPTSRQISLGQQYAPALDNIAMQGL---------KASTVFGEVQSL 442
IT +VP Y ++P IS G + P L+ I Q + + + G Q
Sbjct 462 ITMLVPEPAYSQGLHPDLASISFGDDFNPELNGIGFQLVPRHRFSMMPRGFNLTGLDQQ- 520
Query 443 GATSPTYSNSVFAVPGFKLQDTNYVGYEPAWSELMTAVSKPHGRLCND--LDYWVLSRDY 500
TSP + N+ V +T VG E AWS L T S+ HG + YWVL+R
Sbjct 521 --TSPWFGNAGTGV--VIDPNTVSVGEEVAWSWLRTDYSRLHGDFAQNGNYQYWVLTR-- 574
Query 501 GRNLASVMDTPAYKSFVSAAGAHVEELALQRLTAFFKRIYVSPSSCPYILCGDFNYVFYD 560
+ ++ AG L + Y++P D+ YVF D
Sbjct 575 -----------RFTTYFPDAGTGF------YLDGEYTGTYINPL--------DWQYVFVD 609
Query 561 QRATAENFVLDNVADIVV 578
Q A NF D+ V
Sbjct 610 QTLMAGNFSYYGTFDLKV 627
> Alpavirinae_Human_gut_31_037_Microviridae_AG0297_putative.VP1
Length=668
Score = 58.9 bits (141), Expect = 7e-11, Method: Compositional matrix adjust.
Identities = 56/274 (20%), Positives = 108/274 (39%), Gaps = 31/274 (11%)
Query 250 YFFAGRQSLFQRGFPSYYLEAWLKTSSFT-DAAVD----VSTSGNSVSMRNITFASRMQR 304
+ F+ L + + S + W+ T D ++ V+ ++ + A+++
Sbjct 370 FLFSPEWGLVTKTYQSDLCQNWINTEWLDGDDGINQITAVAVEDGKFTIDALNLANKVYN 429
Query 305 YMDLAFAGGGRNSDFYESQFDVKLNQDNTCPAFLGSDSFDMNANTLYQTTGFEDSSSPLG 364
Y++ G + E+ + + P + G S ++ + T+ D PLG
Sbjct 430 YLNRIAISDGSYRSWLETTWTGSYTERTETPIYCGGSSAEIIFQEVVSTSAATDE--PLG 487
Query 365 AFSGQLSGGTRFRRRNYHFN----DDGYFMEITSIVPRTYYPSYINPTSRQISLGQQYAP 420
+ +G+ GT + + + GY + ITSI PR Y S+ + P
Sbjct 488 SLAGR---GTDSNHKGGYVTVRATEPGYLIGITSITPRLDYTQGNQWDVNLDSIDDLHKP 544
Query 421 ALDNIAMQGLKASTVFGEVQSLGATSPTYSNSVFAVPGFKLQDTNYVGYEPAWSELMTAV 480
ALD I Q L A + + + T + ++G +PAW + MT +
Sbjct 545 ALDGIGFQDLSAELLHAGTTQINTINDTITQK-------------FIGKQPAWIDYMTNI 591
Query 481 SKPHGRLCNDLDYWVLSR----DYGRNLASVMDT 510
++ +G + ++ +LSR DY +N M T
Sbjct 592 NRAYGNFRTNENFMILSRQYSLDYKKNTIKDMTT 625
Score = 35.4 bits (80), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 19/57 (33%), Positives = 27/57 (47%), Gaps = 0/57 (0%)
Query 23 SASWGTLIPTNVTRVVAGDDFSFQPGVGVQALPIVAPFMGSVCVKKEYFFIPDRIYN 79
+ S GTL+P + D F + V P V P GS ++ + FF P R+YN
Sbjct 38 TQSVGTLVPFISIPMCKDDTFKIRLTPNVLTHPTVGPLFGSFKLQNDIFFCPFRLYN 94
> Pichovirinae_Bourget_523_Microviridae_AG0333_putative.VP1
Length=518
Score = 57.8 bits (138), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 49/226 (22%), Positives = 93/226 (41%), Gaps = 31/226 (14%)
Query 279 DAAVDVSTSGNSVSMRNITFASRMQRYMDLAFAGGGRNSDFYESQFDVKLNQDNTC-PAF 337
D + +S + + + ++ A R+Q +++ GG R + + F VK + P +
Sbjct 268 DNSSQLSGTAEAADINSLRRAFRLQEWLERNARGGTRYIESILAHFGVKSSDARLQRPEY 327
Query 338 LGSDSFDMNANTLYQTTGFEDSSSPLGAFSGQ---LSGGTRFRRRNYHFNDDGYFMEITS 394
LG M + + T +++ P+G +G +SGG F+ Y+ + G+ + I S
Sbjct 328 LGGSKGKMVISEVLSTA---ETTLPVGNMAGHGISVSGGNEFK---YNVEEHGWIIGIIS 381
Query 395 IVPRTYYPSYINPTSRQISLGQQYAPALDNIAMQGLKASTVFGEVQSLGATSPTYSNSVF 454
+ P T Y I+ + ++ + P NI Q + ++ ++G T
Sbjct 382 VTPETAYQQGIHRSLSKLDRLDYFWPTFANIGEQEVLGKEIYANGINIGET--------- 432
Query 455 AVPGFKLQDTNYVGYEPAWSELMTAVSKPHGRLCNDLDYWVLSRDY 500
GY P ++E S+ G + LDYW L R +
Sbjct 433 ------------FGYVPRYAEYKFLNSRVAGEMRTSLDYWHLGRKF 466
Score = 43.5 bits (101), Expect = 4e-06, Method: Compositional matrix adjust.
Identities = 31/112 (28%), Positives = 46/112 (41%), Gaps = 0/112 (0%)
Query 27 GTLIPTNVTRVVAGDDFSFQPGVGVQALPIVAPFMGSVCVKKEYFFIPDRIYNVDRQLNF 86
G L PT V V GD ++ P++AP M V V YFF+P+RI + +
Sbjct 33 GGLYPTCVMECVPGDKVKIGTETMLRFAPLIAPVMHKVNVTTHYFFVPNRILWPNWEQWI 92
Query 87 QGVTDTPNTVYKPSIAPPIPFDISKPSGDAISFPVSALQTSVPSGSLGAIVG 138
G D ++ +P K D + +P ++P GA VG
Sbjct 93 TGNLDVQAPWFRLFPSPASGGFHVKSLADYLGYPTLLTPNAIPYPEPGAQVG 144
> Pichovirinae_66_Microbialites_001_Microviridae_AG0132_putative.VP1
Length=518
Score = 55.1 bits (131), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 28/79 (35%), Positives = 42/79 (53%), Gaps = 0/79 (0%)
Query 14 FKLFSGNPTSASWGTLIPTNVTRVVAGDDFSFQPGVGVQALPIVAPFMGSVCVKKEYFFI 73
F L +S G L+PTNV + GD F+ +P ++ LP++AP M V V YFF+
Sbjct 22 FDLSHDVKSSFKMGQLVPTNVIECMPGDLFTIRPQNMLRFLPLIAPVMHRVEVTTHYFFV 81
Query 74 PDRIYNVDRQLNFQGVTDT 92
P+R+ D + G +D
Sbjct 82 PNRLLWADWEKWITGESDV 100
Score = 39.7 bits (91), Expect = 6e-05, Method: Compositional matrix adjust.
Identities = 45/219 (21%), Positives = 84/219 (38%), Gaps = 28/219 (13%)
Query 284 VSTSGNSVSMRNITFASRMQRYMDLAFAGGGRNSDFYESQFDVKLNQDNTC-PAFLGSDS 342
V ++V + + A R+Q +++ G R + S F V+ + P +LG
Sbjct 270 VDVQADAVDINTVRRAFRLQEWLEKNARAGTRYIESILSHFGVRSSDARLQRPEYLGGTK 329
Query 343 FDMNANTLYQTTGFEDSSSPLGAFSGQ---LSGGTRFRRRNYHFNDDGYFMEITSIVPRT 399
+M + + T D++ +G +G GG + Y + G+ + I ++ P T
Sbjct 330 GNMVISEVLATAETTDANVAVGTMAGHGITAHGGNTIK---YRCEEHGWIIGIINVQPVT 386
Query 400 YYPSYINPTSRQISLGQQYAPALDNIAMQGLKASTVFGEVQSLGATSPTYSNSVFAVPGF 459
Y + + P NI Q + ++ A SP + +F
Sbjct 387 AYQQGLPREFSRFDRLDYPWPVFANIGEQEVYNKELY-------ANSPA-PDEIF----- 433
Query 460 KLQDTNYVGYEPAWSELMTAVSKPHGRLCNDLDYWVLSR 498
GY P ++E+ S+ G + + LD+W L R
Sbjct 434 --------GYIPRYAEMKFKNSRVAGEMRDTLDFWHLGR 464
> Alpavirinae_Human_feces_B_007_Microviridae_AG068_putative.VP1
Length=780
Score = 51.2 bits (121), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 54/223 (24%), Positives = 91/223 (41%), Gaps = 29/223 (13%)
Query 286 TSGNSVS-MRNITFASRMQRYMDLAFAGGGRNSDFYESQFDVKLNQDN-TCPAFLGSDSF 343
TSG S++ RN+ + QRY++L G + E +FDV + D P +LG +
Sbjct 525 TSGISINDFRNV---NAYQRYLELNQFRGFSYKEIIEGRFDVNVRYDALNMPEYLGGITR 581
Query 344 DMNANTLYQTTGFEDSSSPLGAFSGQLSGGTRFRRRNYHF----NDDGYFMEITSIVPRT 399
D+ N + QT D+ S +G+ Q T F + +++ M I ++P
Sbjct 582 DIVVNPITQTVETTDTGSYVGSLGSQAGLATCFGNSDGSISVFCDEESIVMGIMYVMPMP 641
Query 400 YYPSYINPTSRQISLGQQYAPALDNIAMQGLKASTVFGEVQSLGATSPTYSNSVFAVPGF 459
Y S + + P D+I Q P Y + A+ F
Sbjct 642 VYDSILPKWLTYRERLDSFNPEFDHIGYQ------------------PIYLKELAAIQAF 683
Query 460 KL-QDTNYV-GYEPAWSELMTAVSKPHGRLCNDLDYWVLSRDY 500
+ +D N V GY+ W E + V + HG + L +++ R +
Sbjct 684 ESGKDLNTVFGYQRPWYEYVQKVDRAHGLFLSSLRNFIMFRSF 726
Lambda K H a alpha
0.318 0.134 0.396 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 54631388