bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: Microviridae_proteins.fasta
575 sequences; 147,441 total letters
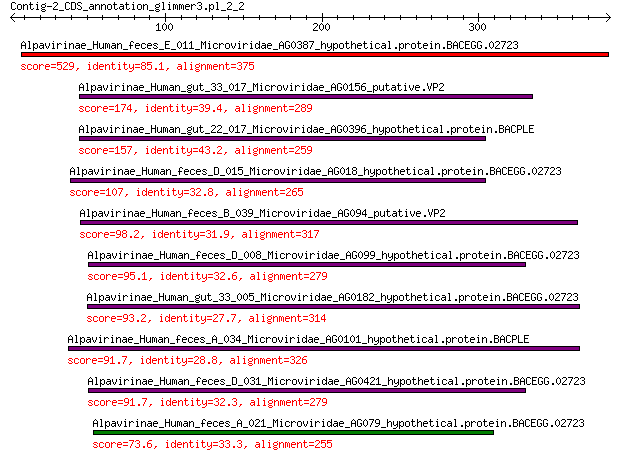
Query= Contig-2_CDS_annotation_glimmer3.pl_2_2
Length=383
Score E
Sequences producing significant alignments: (Bits) Value
Alpavirinae_Human_feces_E_011_Microviridae_AG0387_hypothetical.... 529 0.0
Alpavirinae_Human_gut_33_017_Microviridae_AG0156_putative.VP2 174 5e-52
Alpavirinae_Human_gut_22_017_Microviridae_AG0396_hypothetical.p... 157 2e-45
Alpavirinae_Human_feces_D_015_Microviridae_AG018_hypothetical.p... 107 1e-27
Alpavirinae_Human_feces_B_039_Microviridae_AG094_putative.VP2 98.2 9e-25
Alpavirinae_Human_feces_D_008_Microviridae_AG099_hypothetical.p... 95.1 2e-23
Alpavirinae_Human_gut_33_005_Microviridae_AG0182_hypothetical.p... 93.2 4e-23
Alpavirinae_Human_feces_A_034_Microviridae_AG0101_hypothetical.... 91.7 2e-22
Alpavirinae_Human_feces_D_031_Microviridae_AG0421_hypothetical.... 91.7 3e-22
Alpavirinae_Human_feces_A_021_Microviridae_AG079_hypothetical.p... 73.6 4e-16
> Alpavirinae_Human_feces_E_011_Microviridae_AG0387_hypothetical.protein.BACEGG.02723
Length=410
Score = 529 bits (1362), Expect = 0.0, Method: Compositional matrix adjust.
Identities = 319/375 (85%), Positives = 347/375 (93%), Gaps = 0/375 (0%)
Query 8 IQLNNNSFVQFGIPPlvaaglasgaaslvgNLFGTSSSNSKNMQINKMNNEFNAREAEKA 67
IQLN++ F+ G+ PL A G+ GA S VGN+FG++ SNS+NM+IN+MNNEFNAREAEKA
Sbjct 35 IQLNDSLFILCGLDPLSAIGVGLGAVSGVGNIFGSALSNSQNMKINRMNNEFNAREAEKA 94
Query 68 RQYQTEMWNKTNEWNSPKNIRKRLQEAGYNPYLGMDSSNVGTaqsigssspasaappiqn 127
RQYQ+EMWNKTN+WNSPKN+RKRLQEAGYNPYLG+DSSNVGTAQS GSSSPASAAPPIQN
Sbjct 95 RQYQSEMWNKTNDWNSPKNVRKRLQEAGYNPYLGLDSSNVGTAQSAGSSSPASAAPPIQN 154
Query 128 npMQFDGIQNALSTAIQMDNATNVSNAEVFNLQGQKSLADANAADILSNIDWYKLTPEYR 187
NP+QFDG QNALSTAIQM N+T VSNAE NLQGQK LADA AA LS IDWYK TPEYR
Sbjct 155 NPIQFDGFQNALSTAIQMSNSTKVSNAEANNLQGQKGLADAQAAATLSGIDWYKFTPEYR 214
Query 188 NWLQTTGMSRAQLSFNTDRQNLENMQWTNKIQRAQRTDILLSNEAKRIINKYldssqslq 247
WLQTTGM+RAQLSFNTD+QNLENM+W NKIQRAQRTDILLSN+AK IINKYLD+SQSLQ
Sbjct 215 AWLQTTGMARAQLSFNTDQQNLENMKWVNKIQRAQRTDILLSNQAKGIINKYLDTSQSLQ 274
Query 248 lkllanqsFQAFASGRLSLQQCKTEVTKQLMNMAETEGKKISNKVASETADQLIGALQWQ 307
LKL+ANQSFQAFASGRLSLQQCKTEVTKQLMNMAETEGKKISNK+ASETADQLIGALQWQ
Sbjct 275 LKLMANQSFQAFASGRLSLQQCKTEVTKQLMNMAETEGKKISNKIASETADQLIGALQWQ 334
Query 308 YSSDEMFSRGYAGYAREAGKSRGKGDVAKGKLDEYNYSSRYWNTGIESIGRIGNGIglpl 367
YS+DEM+SRGYAGYAREAGKSRGKGDVAKG+LDEY+YSSRYWNTGIESI RIGNGIGLPL
Sbjct 335 YSADEMYSRGYAGYAREAGKSRGKGDVAKGQLDEYDYSSRYWNTGIESINRIGNGIGLPL 394
Query 368 mlgrglrgpKAIKGF 382
MLGRGLRGPK IKGF
Sbjct 395 MLGRGLRGPKRIKGF 409
> Alpavirinae_Human_gut_33_017_Microviridae_AG0156_putative.VP2
Length=353
Score = 174 bits (441), Expect = 5e-52, Method: Compositional matrix adjust.
Identities = 114/291 (39%), Positives = 171/291 (59%), Gaps = 6/291 (2%)
Query 45 SNSKNMQINKMNNEFNAREAEKARQYQTEMWNKTNEWNSPKNIRKRLQEAGYNPYLGMDS 104
+N N +IN+MNN+FN R A + R +Q MWNK N +N+ R+RL+EAG NPYL M+
Sbjct 31 TNQMNYKINQMNNQFNERMAMQQRDFQENMWNKENTYNTASAQRQRLEEAGLNPYLMMNG 90
Query 105 SNVGTaqsigssspasaappiqnnpMQ--FDGIQNALSTAIQMDNATNVSNAEVFNLQGQ 162
+ G +QS G+ + AS++ P Q F GIQ A+ + Q + V A+V +QGQ
Sbjct 91 GSSGVSQSAGTGASASSSGTAVFQPFQADFSGIQQAIGSVFQ----SQVRQAQVSQMQGQ 146
Query 163 KSLADANAADILSNIDWYKLTPEYRNWLQTTGMSRAQLSFNTDRQNLENMQWTNKIQRAQ 222
++LADA A LS +DW K+T E R +L+ TG++RA+L ++ + Q L+NM + ++ +AQ
Sbjct 147 RNLADAQAMQALSQVDWSKMTKETREYLKATGLARARLGYSKEMQELDNMAFAGRLLQAQ 206
Query 223 RTDILLSNEAKRIINKYldssqslqlkllanqsFQAFASGRLSLQQCKTEVTKQLMNMAE 282
T LL +AK ++N+YLD Q L + A+ + + G L+ Q K + +++ A
Sbjct 207 GTSQLLEADAKTVLNRYLDQQQQADLNVKASVYYNQMSQGHLNYNQAKKVIADEILTYAR 266
Query 283 TEGKKISNKVASETADQLIGALQWQYSSDEMFSRGYAGYAREAGKSRGKGD 333
+G+K+SNKVA TAD LI A S+ F A Y RE +SR D
Sbjct 267 IKGQKLSNKVAEATADSLIRATNAANRSNAEFDLEAAKYNRERARSRSIED 317
> Alpavirinae_Human_gut_22_017_Microviridae_AG0396_hypothetical.protein.BACPLE
Length=386
Score = 157 bits (398), Expect = 2e-45, Method: Compositional matrix adjust.
Identities = 112/261 (43%), Positives = 160/261 (61%), Gaps = 6/261 (2%)
Query 45 SNSKNMQINKMNNEFNAREAEKARQYQTEMWNKTNEWNSPKNIRKRLQEAGYNPYLGMDS 104
+N N +IN+MNN+FN R A + R +Q MWNK N +N+ R+RL+EAG NPYL M+
Sbjct 31 TNQMNYKINQMNNQFNERMAIQQRNWQENMWNKENAYNTASAQRQRLEEAGLNPYLMMNG 90
Query 105 SNVGTaqsigssspasaappiqnnpMQFD--GIQNALSTAIQMDNATNVSNAEVFNLQGQ 162
+ G AQS G+ S AS++ P Q D GI +++ Q + + +E LQG
Sbjct 91 GSAGVAQSAGTGSAASSSGNAVMQPFQADYSGIGSSIGNIFQYE----LMQSEKSQLQGA 146
Query 163 KSLADANAADILSNIDWYKLTPEYRNWLQTTGMSRAQLSFNTDRQNLENMQWTNKIQRAQ 222
+ LADA A + LSNIDW KLT E R +L++TG++RAQL + ++Q +NM T + RAQ
Sbjct 147 RQLADAKAMETLSNIDWGKLTDETRGFLKSTGLARAQLGYAKEQQEADNMAMTGLVLRAQ 206
Query 223 RTDILLSNEAKRIINKYldssqslqlkllanqsFQAFASGRLSLQQCKTEVTKQLMNMAE 282
R+ +LL NEAK I+NKYLD Q L L + A +Q A+G +S + K + ++ + A
Sbjct 207 RSGMLLDNEAKGILNKYLDQHQQLDLSVKAADYYQRMAAGYVSYAEAKKALAEEALAAAR 266
Query 283 TEGKKISNKVASETADQLIGA 303
T G+ ISN+VAS A+ I A
Sbjct 267 TRGQNISNEVASRIAESQIAA 287
> Alpavirinae_Human_feces_D_015_Microviridae_AG018_hypothetical.protein.BACEGG.02723
Length=427
Score = 107 bits (267), Expect = 1e-27, Method: Compositional matrix adjust.
Identities = 87/273 (32%), Positives = 145/273 (53%), Gaps = 11/273 (4%)
Query 39 LFGTSSS-------NSKNMQINKMNNEFNAREAEKARQYQTEMWNKTNEWNSPKNIRKRL 91
+FG+S S N N++IN+MNNEFNA+EAEKAR +Q +MWNK N +N+P R RL
Sbjct 45 VFGSSLSARSQRKANEMNLKINQMNNEFNAKEAEKARAFQLDMWNKENAYNTPAAQRARL 104
Query 92 QEAGYNPYLGMDSSNVGTaqsigssspasaappiqnnpMQFDGI-QNALSTAIQMDNATN 150
+E GYN Y M+ ++ G+A + S+S ASAA F + + + A ++ +
Sbjct 105 EEGGYNAY--MNPADAGSASGMSSTSAASAASSAVMQGTDFSSLGEVGVRLAQELKTFSE 162
Query 151 VSNAEVFNLQGQKSLADANAADILSNIDWYKLTPEYRNWLQTTGMSRAQLSFNTDRQNLE 210
++ N K + + + +W ++PE + +G+ A++ R+
Sbjct 163 KKGLDIRNF-SLKDYLQSQIDKMKGDTNWRNVSPEAIRYNIMSGLEAAKIGMENLREQWA 221
Query 211 NMQWTNKIQRAQRTDILLSNEAKRIINKYldssqslqlkllanqsFQAFASGRLSLQQCK 270
N W+N + RA + LL E+K I+NKYLD Q L + A + G+L + + +
Sbjct 222 NQVWSNNLLRANVANSLLDAESKTILNKYLDQQQQADLNVKAAHYEELINRGQLHVVEAR 281
Query 271 TEVTKQLMNMAETEGKKISNKVASETADQLIGA 303
++++++N A G ISN VA+++A L+ A
Sbjct 282 ELLSREVLNYARARGLNISNWVAAKSAKGLVYA 314
> Alpavirinae_Human_feces_B_039_Microviridae_AG094_putative.VP2
Length=380
Score = 98.2 bits (243), Expect = 9e-25, Method: Compositional matrix adjust.
Identities = 101/343 (29%), Positives = 162/343 (47%), Gaps = 46/343 (13%)
Query 46 NSKNMQINKMNNEFNAREAEKARQYQTEMWNKTNEWNSPKNIRKRLQEAGYNPYLGMDSS 105
N N+++ + N FNA +A+ R +Q +MW N +NSP ++ R G NP++ ++
Sbjct 28 NKVNLRMMREQNAFNAEQAQIQRDWQKQMWGMNNAYNSPSSMISR----GLNPFVQGSAA 83
Query 106 NVGTaqsigssspasaappiqnnpMQ--FDGIQNALSTAIQMDNATNVSNAEVFNLQGQK 163
G+ + A+AAP + F + +L++ Q A+ S E G +
Sbjct 84 MAGSKSPASGGAAATAAPVPSMQAYKPNFSSVFQSLASLAQAK-ASEASAGE----SGSR 138
Query 164 SLADANAADILSNIDWYKLTPEYRN--------WLQTTGMSRAQLSFNTDRQNLENMQWT 215
+ +LS D+Y+ ++N W + TG A L +T+ QNL+N Q+
Sbjct 139 ARQTDTVTPLLS--DYYRGLTNWKNLAIGSSGYWNKETGRVSAALDQSTETQNLKNAQFA 196
Query 216 NKIQRAQRTDILLSNEAKRIINKYldssqslqlkllanqsFQAFASGRLSLQQCKTEVTK 275
+I AQ ILL+++A+RI+NKY+D +Q L + A + G L+ +Q +TE+ +
Sbjct 197 ERISAAQEAQILLNSDAQRIMNKYMDQNQQADLFIKAQTLANLQSQGALTEKQIQTEIQR 256
Query 276 QLMNMAETEGKKISNKVASETADQLIGA------LQWQ---YSSDEMFSRGYAGYAREAG 326
++ AE GKKI N+VASETAD LI A LQ++ Y + R + Y
Sbjct 257 AILASAEASGKKIDNRVASETADSLIKAANASNELQYRDSTYDYKNVKLRKHTEY----- 311
Query 327 KSRGKGDVAKGKLDEYNYS-------SRYWNTGIESIGRIGNG 362
K +A K EY + YW + IG I G
Sbjct 312 ----KTSMANQKAAEYGADLARKQGRTHYWESVARGIGSIAAG 350
> Alpavirinae_Human_feces_D_008_Microviridae_AG099_hypothetical.protein.BACEGG.02723
Length=418
Score = 95.1 bits (235), Expect = 2e-23, Method: Compositional matrix adjust.
Identities = 91/303 (30%), Positives = 152/303 (50%), Gaps = 32/303 (11%)
Query 51 QINKMNNEFNAREAEKARQYQT-----------EMWNKTNEWNSPKNIRKRLQEAGYNPY 99
+IN++NNEFNA EA K R +QT + WN+ N +N P R R++ AG+NPY
Sbjct 40 EINQINNEFNASEALKNRDFQTSEREASQQWNLDQWNRENAYNDPSAQRARMEAAGFNPY 99
Query 100 -LGMDSSNVGTaqsigssspasaappiqnnpMQ-FDG----IQNALSTAIQMDNATNVS- 152
+ +D+ + T+ + S S A + + G QN S Q+ NA +
Sbjct 100 NMNIDAGSASTSGAQSSPGSGSQATASHTPSLPAYTGYAADFQNVASGIAQIGNAVSSGI 159
Query 153 NAEVFNLQGQKSLADANAADILSNI----DWYKLTPEYRNWLQTTGMSRAQLSFNTDRQN 208
+A + + G D ADI+S I +W LT Y+ Q + L + ++
Sbjct 160 DARLTSAYGD----DLMKADIMSKIGGNSEW--LTDVYKLGRQNEAPNL--LGIDLRKKR 211
Query 209 LENMQW-TN-KIQRAQRTDILLSNEAKRIINKYldssqslqlkllanqsFQAFASGRLSL 266
LEN+ TN K+ AQ + L E +RI+NK++ + Q + L +F + +G+LS
Sbjct 212 LENLSTETNIKVALAQGALLGLQAEGQRIVNKFMPAQQQAEFFLKTANAFAQYKAGKLSE 271
Query 267 QQCKTEVTKQLMNMAETEGKKISNKVASETADQLIGALQWQYSSDEMFSRGYAGYAREAG 326
Q KT++ +Q + A+T G+K++N++A AD A+ +Y ++ + G+ A +AG
Sbjct 272 AQVKTQIKQQALLEAQTVGQKLNNRLAERLADYQFKAMAAEYRANAAYYNGFYNDAWQAG 331
Query 327 KSR 329
S+
Sbjct 332 MSK 334
> Alpavirinae_Human_gut_33_005_Microviridae_AG0182_hypothetical.protein.BACEGG.02723
Length=354
Score = 93.2 bits (230), Expect = 4e-23, Method: Compositional matrix adjust.
Identities = 87/317 (27%), Positives = 154/317 (49%), Gaps = 38/317 (12%)
Query 50 MQINKMNNEFNAREAEKARQYQTEMWNKTNEWNSPKNIRKRLQEAGYNPYLGMDSSNVGT 109
+++ ++ NE+ + E++K+R + M++ TNEWNS KN R RL+ AG NPYL M+ + GT
Sbjct 34 LEMQQIQNEWASSESQKSRDFAKSMFDATNEWNSAKNQRARLEAAGLNPYLMMNGGSAGT 93
Query 110 aqsigssspasaappiqnnpMQFDGIQNALSTAIQMDNATNVSNAEVFNLQGQKSLADAN 169
+ G + T Q + + + KS++DA
Sbjct 94 --------------ASSTSASTVSGASGSGGTPYQYTPTNMIGDVASY-ASAMKSMSDAR 138
Query 170 AADILSN-IDWYKLTPEYRNWLQTTGMSRAQLSFNTDRQNLENMQWTNKIQRAQRTDILL 228
+ S+ +D Y + P Y + + G + A F T RQ + AQ+ ++LL
Sbjct 139 KTNTESDLLDKYGV-PTYESQI---GKTMADTYF-TQRQ--------ADVAIAQKANLLL 185
Query 229 SNEAKRIINKYldssqslqlkllanqsFQAFASGRLSLQQCKTEVTKQLMNMAETEGKKI 288
N+A+ ++N YL + +QL++ Q + G +S +Q K + +L A T+G+ I
Sbjct 186 RNDAQEVLNMYLPEEKRIQLQMNGAQYWNMIREGVISEEQAKNLIASRLEIEARTQGQHI 245
Query 289 SNKVASETADQLIGALQWQYSSDEMFSRGYAGYAREAGKSRGKGDVAKGKLDEY--NYSS 346
SNK+A TAD +I A + ++ F+RGY+ ++ + G GK+D + +
Sbjct 246 SNKIAKSTADSIIDATRTAKENEAAFNRGYSQFSNDVG-------FRTGKMDRWLQDPVK 298
Query 347 RYWNTGIESIGRIGNGI 363
W+ GI + G+ +G+
Sbjct 299 ARWDRGINNAGKFIDGL 315
> Alpavirinae_Human_feces_A_034_Microviridae_AG0101_hypothetical.protein.BACPLE
Length=365
Score = 91.7 bits (226), Expect = 2e-22, Method: Compositional matrix adjust.
Identities = 94/330 (28%), Positives = 158/330 (48%), Gaps = 36/330 (11%)
Query 38 NLFGTSSSNSKN----MQINKMNNEFNAREAEKARQYQTEMWNKTNEWNSPKNIRKRLQE 93
N G + SKN +Q+ K+ NE+ + E++K+R + M++ +NEWNS K+ R RL+E
Sbjct 18 NAIGNNRQGSKNRKHQLQMQKIQNEWASSESQKSRDFAKSMFDASNEWNSAKSQRARLEE 77
Query 94 AGYNPYLGMDSSNVGTaqsigssspasaappiqnnpMQFDGIQNALSTAIQMDNATNVSN 153
AG NPYL M+ + GT S++ +S + P Q+ TNV
Sbjct 78 AGLNPYLMMNGGSAGT-AQSTSATASSGSSGSGGMPYQY--------------TPTNVIG 122
Query 154 AEVFNLQGQKSLADANAADILSNIDWYKLTPEYRNWLQTTGMSRAQLSFNTDRQNLENMQ 213
KSL+DA + +++ +Y + + T A F Q
Sbjct 123 DVASYAGAMKSLSDARKSGTEADLLGRYGDSDYSSRIANT---EADTYFK---------Q 170
Query 214 WTNKIQRAQRTDILLSNEAKRIINKYldssqslqlkllanqsFQAFASGRLSLQQCKTEV 273
+ + AQ+ ++LLS EA++++N YL + +QL L Q + G + +Q K +
Sbjct 171 RQSDVATAQKANLLLSAEAQQVMNMYLPQEKQIQLSTLGAQYWNMIRDGSIKEEQAKNLL 230
Query 274 TKQLMNMAETEGKKISNKVASETADQLIGALQWQYSSDEMFSRGYAGYAREAGKSRGKGD 333
+L A T G+ ISNKVA TAD +I A ++ ++RGY+ ++ + G GK D
Sbjct 231 ATRLEIEARTAGQHISNKVARSTADSIIDATNTAKMNEAAYNRGYSQFSNDVGYRTGKMD 290
Query 334 VAKGKLDEYNYSSRYWNTGIESIGRIGNGI 363
+ + +R W+ GI + G+ +G+
Sbjct 291 ----RWSQDPVKAR-WDRGINNAGKFIDGL 315
> Alpavirinae_Human_feces_D_031_Microviridae_AG0421_hypothetical.protein.BACEGG.02723
Length=418
Score = 91.7 bits (226), Expect = 3e-22, Method: Compositional matrix adjust.
Identities = 90/303 (30%), Positives = 150/303 (50%), Gaps = 32/303 (11%)
Query 51 QINKMNNEFNAREAEKARQYQT-----------EMWNKTNEWNSPKNIRKRLQEAGYNPY 99
+IN++NNEFNA EA K R +QT + WN+ N +N P R R++ AG+NPY
Sbjct 40 EINQINNEFNASEALKNRDFQTSEREASQQWNLDQWNRENAYNDPSAQRARMEAAGFNPY 99
Query 100 -LGMDSSNVGTaqsigssspasaappiqnnpMQ-FDG----IQNALSTAIQMDNATNVS- 152
+ +D + T+ + S S+A + + G QN S Q+ NA
Sbjct 100 NMNIDPGSGSTSGAQSSPGSGSSATASHTPSLPAYTGYAADFQNVASGIAQIGNAVASGI 159
Query 153 NAEVFNLQGQKSLADANAADILSNI----DWYKLTPEYRNWLQTTGMSRAQLSFNTDRQN 208
+A + + G D ADI+S I +W LT Y+ Q + L + ++
Sbjct 160 DARLTSAYGD----DLMKADIMSKIGGNSEW--LTDVYKLGRQNEAPNL--LGIDLRKKR 211
Query 209 LENMQW-TN-KIQRAQRTDILLSNEAKRIINKYldssqslqlkllanqsFQAFASGRLSL 266
LEN+ TN K+ AQ + L E +RI+NK++ + Q + L +F + +G+LS
Sbjct 212 LENLSTETNIKVALAQGALLGLQAEGQRIVNKFMPAQQQAEFFLKTANAFAQYKAGKLSE 271
Query 267 QQCKTEVTKQLMNMAETEGKKISNKVASETADQLIGALQWQYSSDEMFSRGYAGYAREAG 326
Q KT++ +Q + A+ G+K++N++A AD A+ +Y ++ + G+ A +AG
Sbjct 272 AQVKTQIKQQALLEAQAAGQKLNNRLAERLADYQFKAMAAEYRANAAYYNGFYNDAWQAG 331
Query 327 KSR 329
S+
Sbjct 332 MSK 334
> Alpavirinae_Human_feces_A_021_Microviridae_AG079_hypothetical.protein.BACEGG.02723
Length=397
Score = 73.6 bits (179), Expect = 4e-16, Method: Compositional matrix adjust.
Identities = 85/280 (30%), Positives = 138/280 (49%), Gaps = 31/280 (11%)
Query 54 KMNNEFNAREAEKARQYQTEMWNKTNEWNSPKNIRKRLQEAGYNPYLGMDSSNVGTaqsi 113
++N F + EA+K R ++ +MWNKTNE+NS N R RL+EAG NPY+ M+ N G A S+
Sbjct 14 ELNRNFQSAEAQKNRDFEVDMWNKTNEYNSATNQRARLEEAGLNPYMMMNGGNAGEAGSV 73
Query 114 gssspasaappiqnnpMQFDGIQ--NALSTAIQM--DNATNVSNAEVFNLQG-------- 161
+ S A P + I N++S AI DN S A N +
Sbjct 74 TAPSTPQGAMPGATGDTTENVISGLNSVSNAIGQFYDNMLTQSRATAQNYENFFNDPSSY 133
Query 162 ---------QKSLA-DANAADILSNIDWYKLTPEYRNWLQTTG---MSRAQLSFNTDRQN 208
K L+ D++ + LS ++ +Y NW Q A S + DRQN
Sbjct 134 GKDQWQAMMMKMLSPDSDNSPFLSKDSASRIFGKYGNWKQAGSAVMFDNAVQSSDLDRQN 193
Query 209 LENMQWTNKIQRAQRTDILLSNEAKRIINKYldssqslqlkllanqsFQAFASGRLSLQQ 268
+N+ TN++ +A + LS++A+++INKYLD +S++L + + +A A G+L+ +Q
Sbjct 194 -KNL--TNQMIQANMIQVNLSSDAQKVINKYLDQQESVKLNIQSALYTEAAARGQLTFEQ 250
Query 269 CKTEVTKQLMNMAETEGKKISNKVASETADQLIGALQWQY 308
+ ++ + NM++ +A E A + QY
Sbjct 251 WQGQL---IQNMSDRLDYNTRKAIADEFIRASCEAYKLQY 287
Lambda K H a alpha
0.313 0.127 0.364 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 33550783