bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: Microviridae_proteins.fasta
575 sequences; 147,441 total letters
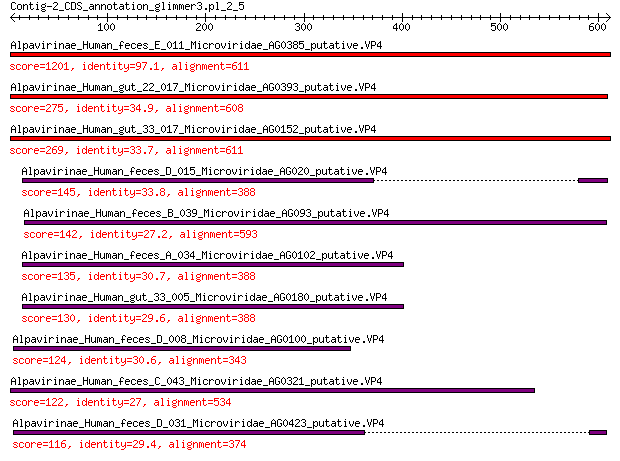
Query= Contig-2_CDS_annotation_glimmer3.pl_2_5
Length=611
Score E
Sequences producing significant alignments: (Bits) Value
Alpavirinae_Human_feces_E_011_Microviridae_AG0385_putative.VP4 1201 0.0
Alpavirinae_Human_gut_22_017_Microviridae_AG0393_putative.VP4 275 3e-85
Alpavirinae_Human_gut_33_017_Microviridae_AG0152_putative.VP4 269 3e-83
Alpavirinae_Human_feces_D_015_Microviridae_AG020_putative.VP4 145 5e-39
Alpavirinae_Human_feces_B_039_Microviridae_AG093_putative.VP4 142 6e-38
Alpavirinae_Human_feces_A_034_Microviridae_AG0102_putative.VP4 135 1e-35
Alpavirinae_Human_gut_33_005_Microviridae_AG0180_putative.VP4 130 5e-34
Alpavirinae_Human_feces_D_008_Microviridae_AG0100_putative.VP4 124 6e-32
Alpavirinae_Human_feces_C_043_Microviridae_AG0321_putative.VP4 122 3e-31
Alpavirinae_Human_feces_D_031_Microviridae_AG0423_putative.VP4 116 3e-29
> Alpavirinae_Human_feces_E_011_Microviridae_AG0385_putative.VP4
Length=611
Score = 1201 bits (3106), Expect = 0.0, Method: Compositional matrix adjust.
Identities = 593/611 (97%), Positives = 601/611 (98%), Gaps = 0/611 (0%)
Query 1 MIPKEELLTKYLFTECLRPQRIVNPYSHDVIFAPCGRCKSCIMNKSNFATAYAMNMATHF 60
MIPKEELLTKYLFTECLRPQRIVNPYSHDVIFAPCG CKSCIMNKSNFATAYAMNMATHF
Sbjct 1 MIPKEELLTKYLFTECLRPQRIVNPYSHDVIFAPCGHCKSCIMNKSNFATAYAMNMATHF 60
Query 61 KYCYFVTLTYKDIFLPYLSVEVVRRSGNRYLFDENFETMVSTSDPRLLTPEYYHDRDLSL 120
KYCYFVTLTYKDIFLPYLSVEVVRRSGNRYLFDENFETMVSTSDPR LTPEYYHDRD SL
Sbjct 61 KYCYFVTLTYKDIFLPYLSVEVVRRSGNRYLFDENFETMVSTSDPRFLTPEYYHDRDFSL 120
Query 121 DPAQNEVEQVFDIGFQSIPRDVSVKSKGSFRFRSFDDEPLKFCIPMKLTELQDILIKANG 180
D AQNEVEQVFD+GFQSIPRDVSVKSKGSFRFRSFDDEPLKFCIPMKLTELQDILIKANG
Sbjct 121 DSAQNEVEQVFDVGFQSIPRDVSVKSKGSFRFRSFDDEPLKFCIPMKLTELQDILIKANG 180
Query 181 RYDYGKKKVVYPSLADCKLQIPVLQSRDIELFFKRLRRNLDSHGFTSSKICYYVVSEYGP 240
RYDYGKKKVVYPSLADCKLQIPVLQSRDIELFFKRLRRNLDSHGFTSSKICYYVVSEYGP
Sbjct 181 RYDYGKKKVVYPSLADCKLQIPVLQSRDIELFFKRLRRNLDSHGFTSSKICYYVVSEYGP 240
Query 241 QTYRPHWHCLLFFNSEEITQTLREDISKAWAYGRIDYSLSRGaaasyvasyvnsaaCLPF 300
QTYRPHWHCLLFFNSEEITQTLREDISKAW+YGRIDYSLSRGAAASYVA+YVNSAACLPF
Sbjct 241 QTYRPHWHCLLFFNSEEITQTLREDISKAWSYGRIDYSLSRGAAASYVAAYVNSAACLPF 300
Query 301 FYVGQKEIRPRSFHSKGFGSNKIFPKSSDVSEISKISDLFFDGVNVDSNGKVINIRPVRS 360
YVGQKEIRPRSFHSKGFGSNK+FPKSSDVSEISKISDLFFDGVNVDSNGKV+NIRPVRS
Sbjct 301 LYVGQKEIRPRSFHSKGFGSNKVFPKSSDVSEISKISDLFFDGVNVDSNGKVVNIRPVRS 360
Query 361 CELAVFPRFSNDFFSDSDTCCKLFQSVIETPERLVSRGYLGIDTPSFGSDGFRLSDLVRA 420
CELAVFPRFSNDFFSDSDTCCKLFQSVIETPERLVSRGYLGIDTP+FGSD FRLSDLVRA
Sbjct 361 CELAVFPRFSNDFFSDSDTCCKLFQSVIETPERLVSRGYLGIDTPNFGSDDFRLSDLVRA 420
Query 421 YSEYYERNFTDFSFSLRFLRGYRTRDYSDELIFREARLFDGYVFNKDMIFGRLYRLFAKV 480
YSEYYERNFTDFSFSLRFLRGYRTRDY+DELIFREARLFDGYVFNKDMIFGRLYRLFAKV
Sbjct 421 YSEYYERNFTDFSFSLRFLRGYRTRDYADELIFREARLFDGYVFNKDMIFGRLYRLFAKV 480
Query 481 LRCFRFWNLKQYTDSWSLKAAVKKIWSYGWEYWKKKEYRFLTTYFEYLEGCNDDERLFLL 540
LRCFRFWNLKQYTDSWSLK A+KKIWSYGWEYWKKKEYRFLTTYFEYLEGCNDDERLFLL
Sbjct 481 LRCFRFWNLKQYTDSWSLKVAIKKIWSYGWEYWKKKEYRFLTTYFEYLEGCNDDERLFLL 540
Query 541 VRTSGSGLATDSPHSWTYTQREDYVNCLPDDLYKRYMKTLKWLTARTEKVLKDKVKHKEF 600
VRTSGSGLATDSPHSWTYTQREDYVNCL DD YKRYM TLKWLTARTEKVLKDKVKHKEF
Sbjct 541 VRTSGSGLATDSPHSWTYTQREDYVNCLSDDFYKRYMNTLKWLTARTEKVLKDKVKHKEF 600
Query 601 NDMQGVLLFSD 611
NDMQGVLLFSD
Sbjct 601 NDMQGVLLFSD 611
> Alpavirinae_Human_gut_22_017_Microviridae_AG0393_putative.VP4
Length=562
Score = 275 bits (703), Expect = 3e-85, Method: Compositional matrix adjust.
Identities = 212/638 (33%), Positives = 299/638 (47%), Gaps = 106/638 (17%)
Query 1 MIPKEELLTKYLFTECLRPQRIVNPYSHDVIFAPCGRCKSCIMNKSNFATAYAMNMATHF 60
MI KE L L T C P+ +VN Y+H+ + CG C SCI+ +S+ T + F
Sbjct 1 MITKE--LQNKLVTRCQHPRTVVNKYTHEPVVVSCGHCPSCILRRSSVQTNLLTTYSAQF 58
Query 61 KYCYFVTLTYKDIFLPYLSVEVVRRSGNRYLFDENFETMVSTSDPRLLTPEYYHDRDLSL 120
+Y YFVTLTY FLP L V VV T D D+S
Sbjct 59 RYVYFVTLTYAPSFLPTLEVSVVE---------------TCTDD----------IADVSC 93
Query 121 DPAQNEVE----QVFDIGFQSIPRDVSVKSKGSFRFRSFDDEPLKFCIPMKLTELQDILI 176
P +E++ + GF+S+PR SVK K S R+F D +KF PMK EL IL
Sbjct 94 VPNIDELDASDPNTYLFGFRSVPRSSSVKLKSSTVERTFKDPDVKFSYPMKPKELLSILG 153
Query 177 KANGRYDYGKKKVVYPSLADCKLQIPVLQSRDIELFFKRLRRNLDSHGFTSSKICYYVVS 236
K N + +IP + +RD++LF KRLR + K+ YY VS
Sbjct 154 KINH---------------NVPNRIPYICNRDLDLFLKRLR-----SYYLDEKLRYYAVS 193
Query 237 EYGPQTYRPHWHCLLFFNSEEITQTLREDISKAWAYGRIDYSLSRGaaasyvasyvnsaa 296
EYGP ++RPHWH LLF NSE ++T+ E++SKAW+YGR D SLSRG AA YVASYVNS
Sbjct 194 EYGPTSFRPHWHLLLFSNSERFSRTVCENVSKAWSYGRCDASLSRGFAAPYVASYVNSFV 253
Query 297 CLPFFYVGQ-KEIRPRSFHSKGFGSNKIFPKSSDVSEISKISDLFFDGVNVDSNGKVINI 355
LP FY K +RP+SFHS GF + +FP+ V+E+ +I+D +GV V+ +G I
Sbjct 254 ALPDFYTQMPKVVRPKSFHSIGFTESNLFPRKVRVAEVDEITDKCLNGVRVERDGYFRTI 313
Query 356 RPVRSCELAVFPRFSNDFFSDSDTCCKLFQSVIETPERLVSRGYLGIDTPSFG-SDGFRL 414
+P L +FPRFS+ + +L + PER++ G I FG S +
Sbjct 314 KPTWPYLLRLFPRFSDAIRKSPSSIYQLLFAAFTAPERVIRSGCADIGCDPFGESSKQSI 373
Query 415 SDLVRAYSEYYERNFTDFSFSLRFLRGYRTRDYSDELIFREARLFDGYVFNKDMIFGRLY 474
+ Y Y + N+ + FL + +SD LI E RL+DG R+Y
Sbjct 374 LSFCKQYLNYVD-NYGKSNEHRNFLSPQASLPHSDVLILSECRLYDGVDLETTHRVSRVY 432
Query 475 RLFAKVLRCFRFWNLKQYTDSWSLKAAVKKIWSYGW--------------------EYWK 514
R F + ++ ++S + WS G +W
Sbjct 433 RF---------FLGISKFIRTYSTDGCSELFWSSGTPGGDLFCRERFLRIISEKIVNFWN 483
Query 515 KKEYRFLTTYFEYLEGCNDDERLFLLVRTSGSGLATDSPHSWTYTQ----REDYVNCLPD 570
+ EY L +++ LE ND E + +R +S+ Y + E N LP
Sbjct 484 RYEYNRLVDFYQTLEDSNDKELVDFELRN----------YSFRYNRSVRDNERPYNELP- 532
Query 571 DLYKRYMKTLKWLTARTEKVLKDKVKHKEFNDMQGVLL 608
++ L A + +DKVKHK+ ND+ G+ L
Sbjct 533 --------LVRRLAAASLMKCRDKVKHKKVNDLSGIFL 562
> Alpavirinae_Human_gut_33_017_Microviridae_AG0152_putative.VP4
Length=565
Score = 269 bits (688), Expect = 3e-83, Method: Compositional matrix adjust.
Identities = 206/628 (33%), Positives = 299/628 (48%), Gaps = 80/628 (13%)
Query 1 MIPKEELLTKYLFTECLRPQRIVNPYSHDVIFAPCGRCKSCIMNKSNFATAYAMNMATHF 60
MI KE L L T C P+ +VN Y+H+ + CG C SC++ +S T + F
Sbjct 1 MITKE--LQNKLVTRCQNPRTVVNKYTHEPVVVSCGACPSCVLRRSGIQTNLLTTYSAQF 58
Query 61 KYCYFVTLTYKDIFLPYLSVEVVRRSGNRYLFDENFETMVSTSDPRLLTPEYYHDRDLSL 120
+Y YFVTLTY FLP L V V+ ++ + S D L
Sbjct 59 RYVYFVTLTYAPCFLPTLEVSVIETCT------DDIADVSSVPDIN------------DL 100
Query 121 DPAQNEVEQVFDIGFQSIPRDVSVKSKGSFRFRSFDDEPLKFCIPMKLTELQDILIKANG 180
DP N + GF S+PR SVK K S R+F D ++F PMK EL IL K N
Sbjct 101 DPCDN---NRYLFGFCSVPRSASVKLKNSTVERTFKDPEVRFSYPMKPKELLSILDKINH 157
Query 181 RYDYGKKKVVYPSLADCKLQIPVLQSRDIELFFKRLRRNLDSHGFTSSKICYYVVSEYGP 240
+ +IP + +RD++LF KRLR + K+ YY VSEYGP
Sbjct 158 ---------------NVPNRIPYICNRDLDLFLKRLR-----SYYPYEKLRYYAVSEYGP 197
Query 241 QTYRPHWHCLLFFNSEEITQTLREDISKAWAYGRIDYSLSRGaaasyvasyvnsaaCLPF 300
+YRPHWH LLF NSE+ ++T+ E++SKAW+YGR D SLSRG AA YVASYVNS LP
Sbjct 198 TSYRPHWHLLLFSNSEQFSKTILENVSKAWSYGRCDASLSRGFAAPYVASYVNSFVALPS 257
Query 301 FYVGQKE-IRPRSFHSKGFGSNKIFPKSSDVSEISKISDLFFDGVNVDSNGKVINIRPVR 359
FY +RP+SFHS GF + +FP+ +SEI +++D +GV V+ +G ++P
Sbjct 258 FYTEMPRFLRPKSFHSIGFTESNLFPRKVRISEIDEVTDKCLNGVRVERDGYFRILKPSW 317
Query 360 SCELAVFPRFSNDFFSDSDTCCKLFQSVIETPERLVSRGYLGIDTPSFGSDGFR-LSDLV 418
L +FPRFS+ + +L + P R++ G I F + + L
Sbjct 318 PYLLRLFPRFSDAIRKSPSSIYQLLFAAFTAPARVIRSGCADIGCDPFVENSKQSLLSFC 377
Query 419 RAYSEYYERNFTDFSFSLRFLRGYRTRDYSDELIFREARLFDGYVFNKDMIFGRLYRLFA 478
+ Y Y + + F FL + +SD LI E RL+DG R+YR F
Sbjct 378 KQYLNYVDNHGKSNEFK-NFLSPHADLPHSDVLILTECRLYDGVDLEAAHRLSRVYRFFL 436
Query 479 KVLRCFRFWNLKQYTDS-WSLKAAVKKIWSYGWE------------YWKKKEYRFLTTYF 525
+ + R ++ ++ WS ++ +G E +W + +Y L ++
Sbjct 437 GIAKFIRTYSTDGCSELFWSSGTPGGEL--FGRERFLRIISEKIVVFWNRYDYMRLVDFY 494
Query 526 EYLEGCNDDERLFLLVRTSGSGLATDSPHSWTYTQREDYVNCLPDDLYKRY--MKTLKWL 583
+ LE ND E + +R +S+ Y + D K Y + ++ L
Sbjct 495 QTLEDSNDKELVDFEIRN----------YSFCYNRSV-------RDKEKPYHELPLVRRL 537
Query 584 TARTEKVLKDKVKHKEFNDMQGVLLFSD 611
A + +DKVKHK+ NDM G+ + D
Sbjct 538 AAASLMKCRDKVKHKKINDMFGIFSYQD 565
> Alpavirinae_Human_feces_D_015_Microviridae_AG020_putative.VP4
Length=563
Score = 145 bits (366), Expect = 5e-39, Method: Compositional matrix adjust.
Identities = 119/384 (31%), Positives = 185/384 (48%), Gaps = 45/384 (12%)
Query 13 FTECLRPQRIVNPYSHDVIFAPCGRCKSCIMNKSNFATAYAMNMATHFKYCYFVTLTYKD 72
F C PQ I N Y+ D + CG+C C++ K++ +T + + +YCYFVTLTY
Sbjct 9 FNRCEHPQVIQNKYTGDYVKVDCGQCPYCLIKKADRSTQKCDFVKYNHRYCYFVTLTYNT 68
Query 73 IFLPYLSVEVVRRSGNRYLFDENFETMVSTSDPRLLT--------PEYY----------- 113
++P +S+ + + +L ++ + R+LT P++
Sbjct 69 QYVPKMSLTQIEDYLSEWLPVRPPKSFGTQLTARMLTDSRVNKKIPDFMSAEVNRPYMLE 128
Query 114 HDRDLSLDPAQ-------NEVEQVFDIGFQSIPRDVSVKSKGSFRFRSFDDEPLKFCIPM 166
H R L D + N + + +SIPR VS + ++F DE + + M
Sbjct 129 HLRLLEADRYKALALRYPNFISKARPYILRSIPR-VS-------KLQNFKDEYFEELVWM 180
Query 167 KLTELQDILIKANGRYDYGKKKVVYPSLADCKLQIPVLQSRDIELFFKRLRRNLDSHGFT 226
L E+ + L K N G + K + + RD +LF KRLR+ L
Sbjct 181 -LPEIAESLKKKNNTDANG-------AFPQFKGLLKYVNIRDYQLFAKRLRKYLSKKVGK 232
Query 227 SSKICYYVVSEYGPQTYRPHWHCLLFFNSEEITQTLREDISKAWAYGRIDYSLSRGaaas 286
KI YVVSEY P+T+RPH+H L FF+S+EI + R+ + ++W GR+D L+R A S
Sbjct 233 YEKIHSYVVSEYSPKTFRPHFHILFFFDSDEIAKNFRQAVYQSWRLGRVDTQLAREQANS 292
Query 287 yvasyvnsaaCLPFFYVGQKEIRPRSFHSKGFGSNKIFPKSSDVSEISKISDLFFDGVNV 346
YV++Y+NS +PF Y +K IRPRS S FG ++ + + FDG++
Sbjct 293 YVSNYLNSVVSIPFVYKAKKSIRPRSRFSNLFGFEEV---KEGIRKAQDKRAALFDGLSY 349
Query 347 DSNGKVINIRPVRSCELAVFPRFS 370
SN K + P S +FPRF+
Sbjct 350 ISNQKFVRYVPSGSLIDRLFPRFT 373
Score = 23.1 bits (48), Expect = 8.1, Method: Compositional matrix adjust.
Identities = 12/30 (40%), Positives = 14/30 (47%), Gaps = 0/30 (0%)
Query 579 TLKWLTARTEKVLKDKVKHKEFNDMQGVLL 608
T KW +V +VKHK ND V L
Sbjct 534 TEKWHDTNYHEVHYFRVKHKVLNDQNDVFL 563
> Alpavirinae_Human_feces_B_039_Microviridae_AG093_putative.VP4
Length=559
Score = 142 bits (357), Expect = 6e-38, Method: Compositional matrix adjust.
Identities = 161/616 (26%), Positives = 253/616 (41%), Gaps = 96/616 (16%)
Query 15 ECLRPQRIVNPYSHDVIFAPCGRCKSCIMNKSNFATAYAMNMATHFKYCYFVTLTYKDIF 74
C I N Y+ I CG+C CI ++ A+ + FKY YFVTLTY +
Sbjct 13 HCQHRSFITNRYNGARITVDCGQCDYCIHKRAQKASMRVKTAGSAFKYSYFVTLTYDNEH 72
Query 75 LPYLSVEVVRRSGNRYLFDENFETMVSTSDPRLLTPEYYHDRDLSLDPAQNEVEQVFDIG 134
+P ++ +V+ +E +V S EY+ +S + ++ + I
Sbjct 73 IPLMNCKVLH---------SEYEDVVGISGDIHFGDEYHKYIPVS-EYRCDDNSMLRHIF 122
Query 135 FQSIPRDVSVKSKGSFRFRSFDDEPLKFCIPMK------LTELQDILIKANGRYDYGKKK 188
F+ + +G+ F D +K +P+K + ++ + K K
Sbjct 123 FEQV--------QGTVPF----DREIKEYVPVKDNWFLSMAAIRSFIYKTQS-----VDK 165
Query 189 VVYPSLADCKLQ--IPVLQSRDIELFFKRLRRNLDSHGFTSSKICYYVVSEYGPQTYRPH 246
YP+ IP L D++ + KRLR+ L +Y V EYGP +RPH
Sbjct 166 TDYPASEQYGRDNLIPFLNYVDVQNYIKRLRKYLFQQLGKYETFHFYAVGEYGPVHFRPH 225
Query 247 WHCLLFFNSEEITQTLREDISKAWAYGRIDYSLSRGaaasyvasyvnsaaCLPFFYVGQK 306
+H LLF NS+++++ LR K+W GR D+ S G A SYVASYVNS P Y +
Sbjct 226 YHLLLFTNSDKVSEVLRYCHDKSWKLGRSDFQRSAGGAGSYVASYVNSLCSAPLLYRSCR 285
Query 307 EIRPRSFHSKGFGSNKIFPKSSDV-------SEISKISDLFFDGVNVDSNGKVINIRPVR 359
RP+S S GF F K D ++I + D +G + NG + P
Sbjct 286 AFRPKSRASVGF-----FEKGCDFVEDDDPYAQIEQKIDSVVNGRVYNFNGVSVRSTPPL 340
Query 360 SCELAVFPRFSNDFFSDSDTCCKLFQSVIETPERLVSRGYLGIDTPSFGSDGFRLSDLVR 419
S + PRFS+ D ++ +V TP+R+ G+ S S LVR
Sbjct 341 SYIRTLLPRFSSARNDDGIAIARILYAVHSTPKRIARFGFADYKQDSILS-------LVR 393
Query 420 AYSEYYERN--FTDFSFSLRFLRGYRTRDYSDELIFREARLFDGYV-----FNKDMIFGR 472
Y +Y + N TD D+LI +R +V + + +
Sbjct 394 TYYQYLKVNPILTD----------------DDKLILHASRCLTRFVNCSSDVDIESYINK 437
Query 473 LYRLFAKVLRCFRFWNLKQY-TDSWSLKAAVKKIWSYGWEYWKKKEYRFLTTYFEYLEGC 531
LYRLF V + FR W+L + +D + + I G EY KKK+Y L F
Sbjct 438 LYRLFLYVYKFFRNWHLPFFGSDISAFSGRIMFIIKTGIEYEKKKDYESLRDVFN----- 492
Query 532 NDDERLFLLVRTSGSGLATDSPHSWTYTQREDYVNCLPDDLYKRYMKTLKWLTARTEKVL 591
+R++ ++ Q D ++ D+ ++ L+ L R+
Sbjct 493 ---------IRSANPNISDCMFALPANGQERDVLS----DVSCETVQLLEQLRLRSATFC 539
Query 592 KDKVKHKEFNDMQGVL 607
+D +KHK ND +
Sbjct 540 RDMIKHKRLNDANNIF 555
> Alpavirinae_Human_feces_A_034_Microviridae_AG0102_putative.VP4
Length=539
Score = 135 bits (339), Expect = 1e-35, Method: Compositional matrix adjust.
Identities = 119/394 (30%), Positives = 180/394 (46%), Gaps = 57/394 (14%)
Query 13 FTECLRPQRIVNPYSHDVIFAPCGRCKSCIMNKSNFATAYAMNMATHFKYCYFVTLTYKD 72
+ CL P+ I N Y+ D I+ PCG C+ C+ NK+ A +YC F+TLTY
Sbjct 9 YGSCLHPRVIKNKYTGDPIYVPCGTCEFCVHNKAIKAELKCNVQLASSRYCEFITLTYST 68
Query 73 IFLPYLSVEVVRRSGNRYLFDENFETMVSTSDPRLLTPEYYHDRDLSLDPAQNEVEQVFD 132
+LP G Y PR +
Sbjct 69 EYLPV---------GKFY------------QGPR------------------------GE 83
Query 133 IGFQSIPRDVSVKSK---GSFRFRSFDDEPLKFCIPMKLTELQDILIKANGRY-DYGKKK 188
+ F S+PRD K G R SF+D+ F + + Q + KA+ Y + +
Sbjct 84 VRFCSLPRDFVYSYKTVQGYTRNISFNDKTFDFDTQLSWSSAQLLQKKAHLHYTSFPDGR 143
Query 189 VVY--PSLADCKLQIPVLQSRDIELFFKRLRRNLDSHGFTSSKICYYVVSEYGPQTYRPH 246
+Y P + + L DI+LFFKRL +N+ T+ KI YYVV EYGP T+RPH
Sbjct 144 CIYNRPYMEGL---VGYLNYHDIQLFFKRLNQNI--RRITNEKIYYYVVGEYGPTTFRPH 198
Query 247 WHCLLFFNSEEITQTLREDISKAWAYGRIDYSLSRGaaasyvasyvnsaaCLPFFYVGQK 306
+H LLF +S ++ +++R+ +SK+W +G D +A+ YVA YVNS ACLP F+ +
Sbjct 199 FHILLFHDSRKLRESIRQFVSKSWRFGDSDTQPVWSSASCYVAGYVNSTACLPDFFKNSR 258
Query 307 EIRPRSFHSKGFGSNKIFPKSSDVSEISKISDLFFDGVNVDSNGKVINIRPVRSCELAVF 366
I+P S F + F + E +I LF+DG ++ NGK +RP RS ++
Sbjct 259 HIKPFGRFSVNFAESA-FNEVFKPEENEEIFSLFYDGRVLNLNGKPTIVRPKRSHINRLY 317
Query 367 PRFSNDFFSDSDTCCKLFQSVIETPERLVSRGYL 400
PR + ++ V P+ + G++
Sbjct 318 PRLDKSKHATVVDDIRIASVVSRLPQVVAKFGFI 351
> Alpavirinae_Human_gut_33_005_Microviridae_AG0180_putative.VP4
Length=536
Score = 130 bits (326), Expect = 5e-34, Method: Compositional matrix adjust.
Identities = 115/394 (29%), Positives = 172/394 (44%), Gaps = 58/394 (15%)
Query 13 FTECLRPQRIVNPYSHDVIFAPCGRCKSCIMNKSNFATAYAMNMATHFKYCYFVTLTYKD 72
+ CL P I N Y+ D I+ CG C+ C+ NK+ + C+FVTLTY
Sbjct 9 YCSCLHPVIIKNKYTGDPIYVECGTCEVCLSNKAIQKELRCNIQLASSRCCFFVTLTYAT 68
Query 73 IFLPYLSVEVVRRSGNRYLFDENFETMVSTSDPRLLTPEYYHDRDLSLDPAQNEVEQVFD 132
+P + S YH
Sbjct 69 EHIPVARFYKLNDS--------------------------YH------------------ 84
Query 133 IGFQSIPRD---VSVKSKGSFRFRSFDDEPLKFCIPMKLTELQDILIKAN---GRYDYGK 186
+PRD V S+G R SF DE + ++ + +L K + Y G+
Sbjct 85 --LCCVPRDHVYTYVTSQGYNRKMSFCDEEFDYPTNLRDEAVTALLDKTHLDRTVYPDGR 142
Query 187 KKVVYPSLADCKLQIPVLQSRDIELFFKRLRRNLDSHGFTSSKICYYVVSEYGPQTYRPH 246
V YP++ D +P L RD++LF KR+ + + +T KI Y V EYGP+T+RPH
Sbjct 143 SVVKYPNMGDL---LPYLNYRDVQLFHKRINQQIKK--YTDEKIYSYTVGEYGPKTFRPH 197
Query 247 WHCLLFFNSEEITQTLREDISKAWAYGRIDYSLSRGaaasyvasyvnsaaCLPFFYVGQK 306
+H L FF+SE + Q + + KAW +G D +A+SYVA Y+NS+ CLP FY +
Sbjct 198 FHLLFFFDSERLAQVFGQLVDKAWRFGNSDTQRVWSSASSYVAGYLNSSHCLPEFYRCNR 257
Query 307 EIRPRSFHSKGFGSNKIFPKSSDVSEISKISDLFFDGVNVDSNGKVINIRPVRSCELAVF 366
+I P S+ F + F ++ E K+ D F +G+ + GK RP RS ++
Sbjct 258 KIAPFGRFSQYFAE-RPFIEAFKPEENEKVFDKFVNGIYLSLGGKPTLCRPKRSLINRLY 316
Query 367 PRFSNDFFSDSDTCCKLFQSVIETPERLVSRGYL 400
P SD D+ + V + P+ L G+L
Sbjct 317 PVLDRSAVSDVDSNVRTALFVSKIPQVLAKFGFL 350
> Alpavirinae_Human_feces_D_008_Microviridae_AG0100_putative.VP4
Length=532
Score = 124 bits (311), Expect = 6e-32, Method: Compositional matrix adjust.
Identities = 105/348 (30%), Positives = 165/348 (47%), Gaps = 49/348 (14%)
Query 4 KEELLTKYLFTECLRPQRIVNPYSHDVIFAPCGRCKSCIMNKSNFATAYAMNMATHFKYC 63
++E + K +F+ C + NPY+ ++I PCG C +C NKS + + ++
Sbjct 3 QQEFIQK-VFSMCQHQVQTKNPYTGELIMVPCGTCPACRFNKSILSQNKVHAQSLISRHV 61
Query 64 YFVTLTYKDIFLPYLSVEVVRRSGNRYLFDENFETMVSTSDPRLLTPEYYHDRDLSLDPA 123
YFVTLTY ++PY E+ D +F + + H RD
Sbjct 62 YFVTLTYAQRYIPYYEYEIE-------ALDADFLAITA------------HCRD------ 96
Query 124 QNEVEQVFDIGFQSIPRDVSVKSKGSFRFRSFDDEPLKFCIPMKLTELQDILIKANGRYD 183
+N + + + ++ + ++ S + +SF + KAN +D
Sbjct 97 RNPMYRTYT--YRGTKHKLRIRGLASPKVKSFS-------FSVNRDYWTSYAQKANLSFD 147
Query 184 YGKKKVVYPSLADCKLQIPVLQSRDIELFFKRLRRNLDSHGFTSSKICYYVVSEYGPQTY 243
GK YP+L+D +IP L D+ L+ KR+R+ + G + I YVV EYGP T+
Sbjct 148 -GK----YPALSD---RIPYLLHDDVSLYMKRVRKYISKLGINET-IHTYVVGEYGPVTF 198
Query 244 RPHWHCLLFFNSEEITQTLREDISKAWAYGRIDYSLSRGaaasyvasyvnsaaCLPFFYV 303
RPH+H LLFFNS+E+ Q++ W +GR+D S SRG A YV+SY+NS + +P
Sbjct 199 RPHFHLLLFFNSDELAQSIVRIARSCWRFGRVDCSASRGDAEDYVSSYLNSFSSIPLHIQ 258
Query 304 GQKEIRPRSFHSKGFG-----SNKIFPKSSDVSEISKISDLFFDGVNV 346
+ IRP + S FG S+ +S D EI L ++G N
Sbjct 259 EIRAIRPFARFSNKFGFSFFESSIKKAQSGDFDEILNGKSLPYNGFNT 306
> Alpavirinae_Human_feces_C_043_Microviridae_AG0321_putative.VP4
Length=546
Score = 122 bits (306), Expect = 3e-31, Method: Compositional matrix adjust.
Identities = 144/548 (26%), Positives = 235/548 (43%), Gaps = 85/548 (16%)
Query 1 MIPKEELLTKYLFTECLRPQRIVNPYSHDVIFAPCGRCKSCIMNKSNFATAYAMNM---A 57
M+P +++ K L CL + + N Y+ +++ CG+C++C+ S A+A ++ + A
Sbjct 1 MLPSDQI-HKELSKTCLHGKNVYNKYTGQMMYQSCGKCEACL---SRLASARSIKVGVQA 56
Query 58 THFKYCYFVTLTYKDIFLPYLSVEVVRRSGNRYLFDENFETMVSTSDPRLLTPEYYHDRD 117
+ KY FVTLTY +P ++ N Y V PR+ YY D
Sbjct 57 SLSKYVMFVTLTYDTYHVP--KCKIYSNGDNNY---------VLVVKPRVKDYFYYETSD 105
Query 118 LSLDPAQNEVEQVFDIGFQSIPRDVSVKSKGSFRFRSFDDEPLKFCIPMKLTELQDILIK 177
Q ++ + S+DD+ F + K + K
Sbjct 106 -----GQKRLKGL-----------------------SYDDD---FRVEFKADKDYIENFK 134
Query 178 ANGRYDYGKKKVVYPSLADCKLQIPVLQSRDIELFFKRLRRNLDSHGFTSSKICYYVVSE 237
D K YP L D L +D +LF KR+R+ + +T KI Y+V E
Sbjct 135 KQANLDV---KGCYPHLKD---MYGYLSRKDCQLFMKRVRKQI--RNYTDEKIHTYIVGE 186
Query 238 YGPQTYRPHWHCLLFFNSEEITQTLREDISKAWAYGRIDYSLSRGaaasyvasyvnsaaC 297
Y P+ +RPH+H L FFNS E++Q+ + W +G +D+S SRG A SYVA YVNS A
Sbjct 187 YSPKHFRPHFHILFFFNSNELSQSFGSIVRSCWKFGLVDWSQSRGDAESYVAGYVNSFAR 246
Query 298 LPFFYVGQKEIRP-----RSFHSKGFGSNKIFPKSSDVSEISKISDLFFDGVNVDSNGKV 352
LP+ +I P F F K + S + + + F +GV NGK+
Sbjct 247 LPYHLGSSPKIAPFGRFSNHFSESCFDDAKQTLRDSFFGQKTPVFAPFLNGVTNLVNGKL 306
Query 353 INIRPVRSCELAVFPRFSNDFFSDSDTCCKLFQS---VIETPERLVSRGYLGIDTPSFGS 409
+ RP RSC + F R + D S L ++ V+E +R +R G++
Sbjct 307 LLTRPSRSCVDSCFFRKARDGRLSSHELLHLIRAVCHVVEQGKRFFARD--GLNYTFLNH 364
Query 410 DGFRLSDLVRAYSEY-YERNFTDFSFSLRF--LRGYRTRDYSDELIFREARLFDGYVFNK 466
F +V+A + Y++ S R L Y D + ++ + D
Sbjct 365 ARF----IVKACKSFRYQKREKLLSLPSRLVTLLYYSRVDLTKDIQDYKNPDVD------ 414
Query 467 DMIFGRLYRLFAKVLRCFRFWNLKQYTDSWSLKAAVKKIWSYGWEYWKKKEYRFLTTYFE 526
+ + +YR+ +R RFW + D + ++ EY++ +Y++L F+
Sbjct 415 EAMAQSIYRVLLYTMRFVRFWKI----DKMPYHEGI-RLLDMCREYYRLLDYQYLRQRFQ 469
Query 527 YLEGCNDD 534
+LE C +D
Sbjct 470 FLEQCEED 477
> Alpavirinae_Human_feces_D_031_Microviridae_AG0423_putative.VP4
Length=532
Score = 116 bits (290), Expect = 3e-29, Method: Compositional matrix adjust.
Identities = 104/357 (29%), Positives = 166/357 (46%), Gaps = 46/357 (13%)
Query 4 KEELLTKYLFTECLRPQRIVNPYSHDVIFAPCGRCKSCIMNKSNFATAYAMNMATHFKYC 63
++E + K +F+ C + NPY+ ++I PCG C +C NKS + + ++
Sbjct 3 QQEFIQK-VFSMCQHQVQTKNPYTGELIMVPCGTCPACRFNKSILSQNKVHAQSLVSRHV 61
Query 64 YFVTLTYKDIFLPYLSVEVVRRSGNRYLFDENFETMVSTSDPRLLTPEYYHDRDLSLDPA 123
YFVTLTY ++PY E+ D +F + + R P Y R +
Sbjct 62 YFVTLTYAQRYIPYYEYEIE-------ALDADFLAITAHCCDR--NPMY---RTYTYRGT 109
Query 124 QNEVEQVFDIGFQSIPRDVSVKSKGSFRFRSFDDEPLKFCIPMKLTELQDILIKANGRYD 183
++++ R ++ + SF F D + Q + NG+Y
Sbjct 110 KHKLR----------IRGLASPNVKSFSFSVNRDYWTSYA--------QKANLSFNGKY- 150
Query 184 YGKKKVVYPSLADCKLQIPVLQSRDIELFFKRLRRNLDSHGFTSSKICYYVVSEYGPQTY 243
P+L+ +IP L D+ L+ KR+R+ + G + I Y+V EYGP ++
Sbjct 151 --------PALSG---RIPYLLHGDVSLYMKRVRKYISKLGINET-IHTYIVGEYGPSSF 198
Query 244 RPHWHCLLFFNSEEITQTLREDISKAWAYGRIDYSLSRGaaasyvasyvnsaaCLPFFYV 303
RPH+H LLFF+S+E+ Q + S W +GR+D S SRG A YV+SY+NS + +P
Sbjct 199 RPHFHLLLFFDSDELAQNIIRIASSCWRFGRVDCSASRGDAEDYVSSYLNSFSSIPLHIQ 258
Query 304 GQKEIRPRSFHSKGFGSNKIFPKSSDVSEISKISDLFFDGVNVDSNGKVINIRPVRS 360
+ IRP + S FG + F +SS S D +G ++ NG I P R+
Sbjct 259 EIRAIRPFARFSNKFGYS--FFESSIKKAQSGNFDEILNGKSLPYNGFDTTIFPWRA 313
Score = 22.7 bits (47), Expect = 9.2, Method: Compositional matrix adjust.
Identities = 6/17 (35%), Positives = 12/17 (71%), Gaps = 0/17 (0%)
Query 591 LKDKVKHKEFNDMQGVL 607
++ ++KH+E ND G+
Sbjct 505 IRKRIKHREINDAVGIF 521
Lambda K H a alpha
0.325 0.140 0.439 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 56326467