bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: Microviridae_proteins.fasta
575 sequences; 147,441 total letters
Query= Contig-35_CDS_annotation_glimmer3.pl_2_2
Length=605
Score E
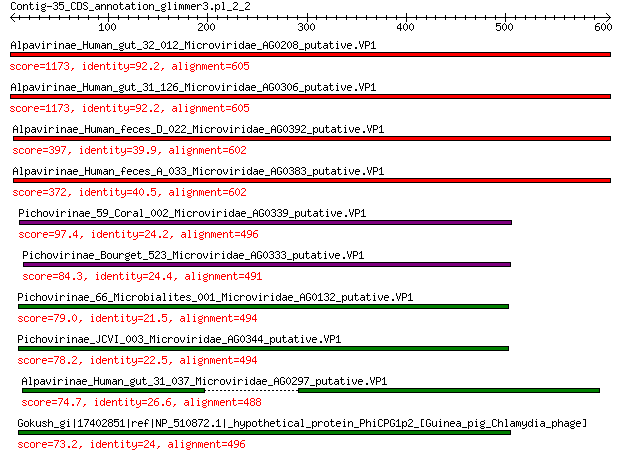
Sequences producing significant alignments: (Bits) Value
Alpavirinae_Human_gut_32_012_Microviridae_AG0208_putative.VP1 1173 0.0
Alpavirinae_Human_gut_31_126_Microviridae_AG0306_putative.VP1 1173 0.0
Alpavirinae_Human_feces_D_022_Microviridae_AG0392_putative.VP1 397 1e-131
Alpavirinae_Human_feces_A_033_Microviridae_AG0383_putative.VP1 372 5e-122
Pichovirinae_59_Coral_002_Microviridae_AG0339_putative.VP1 97.4 4e-23
Pichovirinae_Bourget_523_Microviridae_AG0333_putative.VP1 84.3 5e-19
Pichovirinae_66_Microbialites_001_Microviridae_AG0132_putative.VP1 79.0 2e-17
Pichovirinae_JCVI_003_Microviridae_AG0344_putative.VP1 78.2 5e-17
Alpavirinae_Human_gut_31_037_Microviridae_AG0297_putative.VP1 74.7 8e-16
Gokush_gi|17402851|ref|NP_510872.1|_hypothetical_protein_PhiCPG... 73.2 2e-15
> Alpavirinae_Human_gut_32_012_Microviridae_AG0208_putative.VP1
Length=604
Score = 1173 bits (3034), Expect = 0.0, Method: Compositional matrix adjust.
Identities = 558/605 (92%), Positives = 587/605 (97%), Gaps = 1/605 (0%)
Query 1 MGMMFLTRSRNKKSNVNLSHVSPTTLAPGNLIPISFTRIFAGDDLRFKPSAFVQAMPMNA 60
MGMMFLTRSRNKKSNVNLSHVSPTT+APGNL+PISFTRIFAGDDLRF+PSAFVQAMPMNA
Sbjct 1 MGMMFLTRSRNKKSNVNLSHVSPTTIAPGNLVPISFTRIFAGDDLRFEPSAFVQAMPMNA 60
Query 61 PLVNGFKLCLEYFFVPDRLYNWELLMDNTGVTDDPDKVKFPQISSPAEYSTGTIKFSLNS 120
PLVNGFKLCLEYFFVPDRLYNW LLMDNTGVTD+PD VKFPQI +PAEY++ TIKFSLNS
Sbjct 61 PLVNGFKLCLEYFFVPDRLYNWNLLMDNTGVTDNPDSVKFPQIHAPAEYTSDTIKFSLNS 120
Query 121 QSAARGDAARLANSIVQPGSLADYCGFPVGLFPTYDIVSDTDDRNQFCALKLLGVLDIFY 180
+SAA+G+AARLANSIVQPGSLADYCGFPVGLFPTY++V DTDDRNQFCALKLLGVLDIFY
Sbjct 121 ESAAKGNAARLANSIVQPGSLADYCGFPVGLFPTYEVVLDTDDRNQFCALKLLGVLDIFY 180
Query 181 HYYVNQQIERFPTASFTPTLNSGSEDERNVDYPVTMLRSFLDFVKRSPNPASAIGEWATS 240
HYYVNQQI+RFPTASFTPTLN GS++ERNVDY VTMLRSFLDFVKRS +PA AIG+WATS
Sbjct 181 HYYVNQQIDRFPTASFTPTLNHGSDEERNVDYSVTMLRSFLDFVKRSADPAGAIGQWATS 240
Query 241 NPNGNPIFGTWSWFCSRASIFQRCLPPYYLESWLATSGYEDSEIKVDLDADGKSISFRNI 300
NPNGNPIFGTWSWFCSRASIFQRCLPPYYLESWLATSGYEDSEIKVDL ADGKSISFRNI
Sbjct 241 NPNGNPIFGTWSWFCSRASIFQRCLPPYYLESWLATSGYEDSEIKVDLGADGKSISFRNI 300
Query 301 AAHSHIQRWLDLALAGGSRYSDYINSQFDVSRLKHTTSPLYLGSDRQYLGSNVIYQTTGA 360
AA SHIQRWLDLALAGGSRYSDYINSQFDVSRLKHTTSPLYLGSDRQYLGSNVIYQTTGA
Sbjct 301 AAQSHIQRWLDLALAGGSRYSDYINSQFDVSRLKHTTSPLYLGSDRQYLGSNVIYQTTGA 360
Query 361 GDSSSPLGAFAGQASGGETFRQRSYHFGENGYFVVMASLVPDVIYSRGMDPFNREKTLGD 420
GDS+SPLGAFAGQ+SGGETFR+RSYHFGENGYFVVMASLVPDV+YSRGMDPFNREKTLGD
Sbjct 361 GDSASPLGAFAGQSSGGETFRRRSYHFGENGYFVVMASLVPDVVYSRGMDPFNREKTLGD 420
Query 421 VYVPALDNIAMEPLMVEQVDALPSLLSLQRGTDSVYTLSFRPDKLIKNSALGYVPAWSKV 480
VYVPALDNIAMEPLM+EQVDA+PSL SLQ+ DSVYTLS RPDKLIKNSALGYVPAWSKV
Sbjct 421 VYVPALDNIAMEPLMIEQVDAIPSLRSLQK-VDSVYTLSIRPDKLIKNSALGYVPAWSKV 479
Query 481 MQSTSRAHGRLTTDLKYWLLNRDYGVDIDQIKNTGVIGVDVLKNLASELQQAYQAGYISF 540
MQ+ SRAHGRLTTDLKYWLL+RDYGVD+D+IKNTGVIG DVLKNLASEL QAYQAGYISF
Sbjct 480 MQNVSRAHGRLTTDLKYWLLSRDYGVDVDKIKNTGVIGADVLKNLASELNQAYQAGYISF 539
Query 541 EQMDALVTLFSRIQALPEYTPYVLPNAYNDVFADTSNQAQNFVLTCTFSMSCNREKGKVN 600
EQMDALVTLFSRIQALPEYTPYVLPNAYNDVFADTSNQAQNFVLTCTFSM+CNREKGKVN
Sbjct 540 EQMDALVTLFSRIQALPEYTPYVLPNAYNDVFADTSNQAQNFVLTCTFSMTCNREKGKVN 599
Query 601 TPTTI 605
TPTTI
Sbjct 600 TPTTI 604
> Alpavirinae_Human_gut_31_126_Microviridae_AG0306_putative.VP1
Length=604
Score = 1173 bits (3034), Expect = 0.0, Method: Compositional matrix adjust.
Identities = 558/605 (92%), Positives = 587/605 (97%), Gaps = 1/605 (0%)
Query 1 MGMMFLTRSRNKKSNVNLSHVSPTTLAPGNLIPISFTRIFAGDDLRFKPSAFVQAMPMNA 60
MGMMFLTRSRNKKSNVNLSHVSPTT+APGNL+PISFTRIFAGDDLRF+PSAFVQAMPMNA
Sbjct 1 MGMMFLTRSRNKKSNVNLSHVSPTTIAPGNLVPISFTRIFAGDDLRFEPSAFVQAMPMNA 60
Query 61 PLVNGFKLCLEYFFVPDRLYNWELLMDNTGVTDDPDKVKFPQISSPAEYSTGTIKFSLNS 120
PLVNGFKLCLEYFFVPDRLYNW LLMDNTGVTD+PD VKFPQI +PAEY++ TIKFSLNS
Sbjct 61 PLVNGFKLCLEYFFVPDRLYNWNLLMDNTGVTDNPDSVKFPQIHAPAEYTSDTIKFSLNS 120
Query 121 QSAARGDAARLANSIVQPGSLADYCGFPVGLFPTYDIVSDTDDRNQFCALKLLGVLDIFY 180
+SAA+G+AARLANSIVQPGSLADYCGFPVGLFPTY++V DTDDRNQFCALKLLGVLDIFY
Sbjct 121 ESAAKGNAARLANSIVQPGSLADYCGFPVGLFPTYEVVLDTDDRNQFCALKLLGVLDIFY 180
Query 181 HYYVNQQIERFPTASFTPTLNSGSEDERNVDYPVTMLRSFLDFVKRSPNPASAIGEWATS 240
HYYVNQQI+RFPTASFTPTLN GS++ERNVDY VTMLRSFLDFVKRS +PA AIG+WATS
Sbjct 181 HYYVNQQIDRFPTASFTPTLNHGSDEERNVDYSVTMLRSFLDFVKRSADPAGAIGQWATS 240
Query 241 NPNGNPIFGTWSWFCSRASIFQRCLPPYYLESWLATSGYEDSEIKVDLDADGKSISFRNI 300
NPNGNPIFGTWSWFCSRASIFQRCLPPYYLESWLATSGYEDSEIKVDL ADGKSISFRNI
Sbjct 241 NPNGNPIFGTWSWFCSRASIFQRCLPPYYLESWLATSGYEDSEIKVDLGADGKSISFRNI 300
Query 301 AAHSHIQRWLDLALAGGSRYSDYINSQFDVSRLKHTTSPLYLGSDRQYLGSNVIYQTTGA 360
AA SHIQRWLDLALAGGSRYSDYINSQFDVSRLKHTTSPLYLGSDRQYLGSNVIYQTTGA
Sbjct 301 AAQSHIQRWLDLALAGGSRYSDYINSQFDVSRLKHTTSPLYLGSDRQYLGSNVIYQTTGA 360
Query 361 GDSSSPLGAFAGQASGGETFRQRSYHFGENGYFVVMASLVPDVIYSRGMDPFNREKTLGD 420
GDS+SPLGAFAGQ+SGGETFR+RSYHFGENGYFVVMASLVPDV+YSRGMDPFNREKTLGD
Sbjct 361 GDSASPLGAFAGQSSGGETFRRRSYHFGENGYFVVMASLVPDVVYSRGMDPFNREKTLGD 420
Query 421 VYVPALDNIAMEPLMVEQVDALPSLLSLQRGTDSVYTLSFRPDKLIKNSALGYVPAWSKV 480
VYVPALDNIAMEPLM+EQVDA+PSL SLQ+ DSVYTLS RPDKLIKNSALGYVPAWSKV
Sbjct 421 VYVPALDNIAMEPLMIEQVDAIPSLRSLQK-VDSVYTLSIRPDKLIKNSALGYVPAWSKV 479
Query 481 MQSTSRAHGRLTTDLKYWLLNRDYGVDIDQIKNTGVIGVDVLKNLASELQQAYQAGYISF 540
MQ+ SRAHGRLTTDLKYWLL+RDYGVD+D+IKNTGVIG DVLKNLASEL QAYQAGYISF
Sbjct 480 MQNVSRAHGRLTTDLKYWLLSRDYGVDVDKIKNTGVIGADVLKNLASELNQAYQAGYISF 539
Query 541 EQMDALVTLFSRIQALPEYTPYVLPNAYNDVFADTSNQAQNFVLTCTFSMSCNREKGKVN 600
EQMDALVTLFSRIQALPEYTPYVLPNAYNDVFADTSNQAQNFVLTCTFSM+CNREKGKVN
Sbjct 540 EQMDALVTLFSRIQALPEYTPYVLPNAYNDVFADTSNQAQNFVLTCTFSMTCNREKGKVN 599
Query 601 TPTTI 605
TPTTI
Sbjct 600 TPTTI 604
> Alpavirinae_Human_feces_D_022_Microviridae_AG0392_putative.VP1
Length=589
Score = 397 bits (1019), Expect = 1e-131, Method: Compositional matrix adjust.
Identities = 240/617 (39%), Positives = 343/617 (56%), Gaps = 43/617 (7%)
Query 4 MFLTRSRNKKSNVNLSHVSPTTLAPGNLIPISFTRIFAGDDLRFKPSAFVQAMPMNAPLV 63
MFL+R RNKKS L +PT+ + G LIP + TR+ AGDD F+P VQA+P+ AP +
Sbjct 1 MFLSRKRNKKSRFKLFSGNPTSASWGTLIPTNVTRVIAGDDFSFQPGVGVQALPIVAPFM 60
Query 64 NGFKLCLEYFFVPDRLYNWELLMDNTGVTDDPDKVKFPQISSPAEY--STGTIKFSLNSQ 121
+ EYFF+PDR+YN + ++ GVTD P+ V P ++ P + S + K ++
Sbjct 61 GNVCVKKEYFFIPDRIYNIDRQLNFQGVTDTPNTVYKPSMAPPIPFDISAPSGKVDISVS 120
Query 122 SAARGDAARLANSIVQPGSLADYCGFPVGLFPTYDIVSDTDDRNQFCALKLLGVLDIFYH 181
R IV PGSLADY G G IV+ D + +G +DI+Y+
Sbjct 121 DVQTDLPLRSLGYIVGPGSLADYMGEAPG-----SIVTGVIDLTPY-----IGYIDIYYN 170
Query 182 YYVNQQIERFPTASFTPTLNSGSEDERNVDYP----VTMLRSFLDFVKRSPNPASAIGE- 236
YY+NQQ PT +G+ + ++YP V+ L +L +K +PN + AI E
Sbjct 171 YYLNQQF------GLVPTSLAGTVSDSAMEYPYFLDVSELEGYLRAIKTTPNTSPAIRED 224
Query 237 -WATSNPNGNPIFGT-------WSWFCSRASIFQRCLPPYYLESWLATSGYEDSEIKVDL 288
A+ + N N W +F R S+FQR P YYLE+WL TS + D+ VD+
Sbjct 225 DSASYSTNVNAALRAVASNAFLWDFFTGRQSLFQRGFPSYYLEAWLKTSSFSDA--AVDI 282
Query 289 DADGKSISFRNIAAHSHIQRWLDLALAGGSRYSDYINSQFDVSRLKHTTSPLYLGSDRQY 348
G S+S RNI S +QR++DLA AGG R SD+ +QFDV + T P +LGSD
Sbjct 283 STSGSSVSMRNITFASRMQRYMDLAFAGGGRNSDFYEAQFDVKLNQDNTCPAFLGSDSFD 342
Query 349 LGSNVIYQTTGAGDSSSPLGAFAGQASGGETFRQRSYHFGENGYFVVMASLVPDVIYSRG 408
+ N +YQTTG D+SSPLGAF+GQ SGG FR+R+YHF ++GYF+ + S+VP V Y
Sbjct 343 MNVNTLYQTTGFEDNSSPLGAFSGQLSGGTRFRRRNYHFNDDGYFMEITSIVPRVYYPSY 402
Query 409 MDPFNREKTLGDVYVPALDNIAMEPLMVEQVDALPSLLSLQRGTDSVYTLSFRPDKLIKN 468
++P +R+ +LG Y PALDNIAM+ L V LS T + + LS K+ +
Sbjct 403 INPTSRQISLGQQYAPALDNIAMQGLKASTVFGEVQNLSAISTTYANFILSIPGFKVQDS 462
Query 469 SALGYVPAWSKVMQSTSRAHGRLTTDLKYWLLNRDYGVDIDQIKNTGVIGVDVLKNLASE 528
+ +GY PAWS++M + S+ HGRL DL YW+L+RDYG ++ + ++ G D + +
Sbjct 463 NYVGYEPAWSELMTAVSKPHGRLCNDLDYWVLSRDYGRNLSYVMDSKAYG-DFVSAAGTN 521
Query 529 LQQAYQAGYISFEQMDALVTLFSRIQALPEYTPYVLPNAYNDVFADTSNQAQNFVLTCTF 588
+ + +S +++ A F RI P PY+L +N VF D A+NFVL
Sbjct 522 IDE------LSLQRLTA---FFKRIYESPSSCPYILCGDFNYVFYDQRPTAENFVLDNVA 572
Query 589 SMSCNREKGKVNTPTTI 605
+ REK KVN TT+
Sbjct 573 DIVVFREKSKVNVATTL 589
> Alpavirinae_Human_feces_A_033_Microviridae_AG0383_putative.VP1
Length=590
Score = 372 bits (954), Expect = 5e-122, Method: Compositional matrix adjust.
Identities = 244/628 (39%), Positives = 346/628 (55%), Gaps = 64/628 (10%)
Query 4 MFLTRSRNKKSNVNLSHVSPTTLAPGNLIPISFTRIFAGDDLRFKPSAFVQAMPMNAPLV 63
MFL+R RNKKS L +PT+ + G LIP + TR+ AGDD F+P VQA+P+ AP +
Sbjct 1 MFLSRKRNKKSRFKLFSGNPTSASWGTLIPTNVTRVVAGDDFSFQPGVGVQALPIVAPFM 60
Query 64 NGFKLCLEYFFVPDRLYNWELLMDNTGVTDDPDKVKFPQISSPAEYSTGTIKFSLNSQSA 123
+ EYFF+PDR+YN E ++ GVTD P+ V P ++ P I F + S
Sbjct 61 GSVCVKKEYFFIPDRIYNIERQLNFQGVTDTPNTVYKPSMAPP-------IPFDI---SV 110
Query 124 ARGDAARLANS-------------IVQPGSLADYCGFPVGLFPTYDIVSDTDDRNQFCAL 170
A G+ + S IV PGSLADY G G IV+ D +
Sbjct 111 ASGEEVSIPVSDVQTELPLVSLGDIVGPGSLADYMGEAPG-----SIVTGVIDLTPY--- 162
Query 171 KLLGVLDIFYHYYVNQQIERFPTASFTPTLNSGSEDERNVDYP----VTMLRSFLDFVKR 226
+G +DI+Y+YYVNQQ + PT+ +G++ + V+YP V+ L ++L +K
Sbjct 163 --IGYIDIYYNYYVNQQYDLVPTSL------AGTKSDSAVEYPYYLSVSELETYLRTIKT 214
Query 227 SPNPASAIG-----EWATSNPNGNPIFGT----WSWFCSRASIFQRCLPPYYLESWLATS 277
+PN + AI ++T+ G+ W++F R S+FQR P YYLE+WL TS
Sbjct 215 TPNTSPAIRVDDSVSYSTNVEKALQAVGSYAFSWNFFTGRQSLFQRGFPSYYLEAWLKTS 274
Query 278 GYEDSEIKVDLDADGKSISFRNIAAHSHIQRWLDLALAGGSRYSDYINSQFDVSRLKHTT 337
+ D+ VD+ A G S+S RNI S +QR++DLA AGG R SD+ SQFDV + T
Sbjct 275 SFVDA--AVDVSASGSSVSMRNITFASRMQRYMDLAFAGGGRNSDFYESQFDVKLDQDNT 332
Query 338 SPLYLGSDRQYLGSNVIYQTTGAGDSSSPLGAFAGQASGGETFRQRSYHFGENGYFVVMA 397
P +LGSD + N +YQTTG D+SSPLGAF+GQ SGG FR+R+YHF ++GYF+ +
Sbjct 333 CPAFLGSDSFDMNVNTLYQTTGFEDNSSPLGAFSGQLSGGTRFRRRNYHFNDDGYFMEIT 392
Query 398 SLVPDVIYSRGMDPFNREKTLGDVYVPALDNIAMEPLMVEQVDALPSLLSLQRGTDSVYT 457
S+VP V Y ++P +R+ +LG Y PALDNIAM+ L V L + +
Sbjct 393 SIVPRVYYPSYINPTSRQISLGQQYAPALDNIAMQGLKASTVFGEVQSLGATNPSYAENR 452
Query 458 LSFRPDKLIKNSALGYVPAWSKVMQSTSRAHGRLTTDLKYWLLNRDYGVDIDQIKNTGVI 517
L KL + +GY PAWS++M + S+ HGRL DL YW+L+RDYG +I + ++
Sbjct 453 LIIPGFKLQDFNYIGYEPAWSELMTAVSKPHGRLCNDLDYWVLSRDYGRNIAHVMDSKAY 512
Query 518 GVDVLKNLASELQQAYQAGYISFEQMDALVTLFSRIQALPEYTPYVLPNAYNDVFADTSN 577
D + S + + +S +++ A F RI P PY+L +N VF D
Sbjct 513 K-DFIAAAGSGVDE------LSLQRLTA---FFKRIYISPSSCPYILCGDFNYVFYDQRP 562
Query 578 QAQNFVLTCTFSMSCNREKGKVNTPTTI 605
A+NF+L + REK KVN TT+
Sbjct 563 TAENFILDNVADIVVFREKSKVNVATTL 590
> Pichovirinae_59_Coral_002_Microviridae_AG0339_putative.VP1
Length=522
Score = 97.4 bits (241), Expect = 4e-23, Method: Compositional matrix adjust.
Identities = 120/529 (23%), Positives = 197/529 (37%), Gaps = 102/529 (19%)
Query 10 RNKKSNVNLSHVSPTTLAPGNLIPISFTRIFAGDDLRFKPSAFVQAMPMNAPLVNGFKLC 69
R + + +LSH + G L+PI+ GD K + + P+ P+++ +
Sbjct 11 RPQTNTFDLSHDRKFSGKIGELMPITVMEAVPGDKFNIKATNLTRFAPLITPIMHQASVY 70
Query 70 LEYFFVPDRLY--NWELLMDNTGVTDDPDKVKFPQISSPAEYSTGTIKFSLNSQSAARGD 127
+FFVP+R+ NWE + +G D FP I N + G
Sbjct 71 CHFFFVPNRILWSNWEDFI--SGGEDGLADPTFPTI---------------NITTPTNGY 113
Query 128 AARLANSIVQPGSLADYCGFPVGLFPTYDIVSDTDDRNQFCALKLLGVLDIFYHYYVNQQ 187
A G+LADY G P G D VS AL I+ YY ++
Sbjct 114 AV---------GALADYLGLPTGT--QIDGVS---------ALPFAAYQKIYDDYYRDEN 153
Query 188 IERFPTASFTPTLNSGSE-----DERNVDYPVTMLRSFLDFVKRSPNPASAIGEWATSNP 242
+ + + SG++ R + S L + +R P +G A
Sbjct 154 LIDKVDITLSDGTQSGADAIELGTMRKRAWQHDYFTSALPWTQRGPEATIPLGTSA---- 209
Query 243 NGNPIFGTWSWFCSRASIFQRC-----LPPYYLE--SWLATSG----YEDSEIKVDLDAD 291
PI TWS + A+I + + Y + S L T G D V LD D
Sbjct 210 ---PI--TWSNNTATATIVRNNTSGNPIAGYTFDGASALYTGGSGQLIADLPTPVSLDFD 264
Query 292 GKSISFRNIAAHS-----------HIQRWLDLALAGGSRYSDYINSQFDV----SRLKHT 336
F ++A + +Q WL+ GG+RY + I + F V +RL+
Sbjct 265 NSDHLFADLAGATASSINDLRRAFRLQEWLERNARGGARYIEIIMAHFGVRSSDARLQR- 323
Query 337 TSPLYLGSDRQYLGSNVIYQTTGAGDSSSPLGAFAGQASGGETFRQRSYHFGENGYFVVM 396
P +LG + + + QT+ +P G AG + SY E+GY + +
Sbjct 324 --PEFLGGSATPITISEVLQTSANNTEPTPQGNMAGHGVSVGSSNYVSYKCEEHGYIIGI 381
Query 397 ASLVPDVIYSRGMDPFNREKTLGDVYVPALDNIAMEPLMVEQVDALPSLLSLQRGTDSVY 456
S++P Y +G+ ++ D Y P+ NI +P++ E++
Sbjct 382 MSVMPKTAYQQGIPKHFKKLDKFDYYWPSFANIGEQPILNEEL----------------- 424
Query 457 TLSFRPDKLIKNSALGYVPAWSKVMQSTSRAHGRLTTDLKYWLLNRDYG 505
+ + GY P +++ S HG L +W + R +G
Sbjct 425 ---YHQNNATDTETFGYTPRYAEYKYIPSTVHGEFRDSLDFWHMGRIFG 470
> Pichovirinae_Bourget_523_Microviridae_AG0333_putative.VP1
Length=518
Score = 84.3 bits (207), Expect = 5e-19, Method: Compositional matrix adjust.
Identities = 120/520 (23%), Positives = 204/520 (39%), Gaps = 99/520 (19%)
Query 14 SNV-NLSHVSPTTLAPGNLIPISFTRIFAGDDLRFKPSAFVQAMPMNAPLVNGFKLCLEY 72
SNV +LSH + G L P GD ++ ++ P+ AP+++ + Y
Sbjct 17 SNVFDLSHDVKMSFKMGGLYPTCVMECVPGDKVKIGTETMLRFAPLIAPVMHKVNVTTHY 76
Query 73 FFVPDRLY--NWELLMDNTGVTDDPDKVKFPQISSPAEYSTGTIKFSLNSQSAARGDAAR 130
FFVP+R+ NWE + TG D V+ P F L A+ G +
Sbjct 77 FFVPNRILWPNWEQWI--TGNLD----VQAPW-------------FRLFPSPASGGFHVK 117
Query 131 LANSIVQPGSLADYCGFPVGLFPTYDIVSDTDDRNQFCALKLLGVLDIFYHYYVNQQIER 190
SLADY G+P L P I + I+ YY +Q ++
Sbjct 118 ---------SLADYLGYPTLLTPN-AIPYPEPGAQVGSPFPVAAYNKIYNEYYRDQNLQ- 166
Query 191 FPTASFTPTLNSGSEDE----RNVDYPVTMLRSFLDFVKRSPNPASAIGEWATSNPNGNP 246
+P L+S ++ E RN+ R++ S P + G+ A + P G+
Sbjct 167 ------SPVLDSLTDGENDPFRNLAAGPVKKRAWQHDYFTSCLPWAQKGD-AVTIPIGDV 219
Query 247 IFGTWSWFCSRASIFQRCL--PPYYLESWLATS---------GYEDSEIKVD----LDAD 291
T ++ + R + PY ++ L S G + +D L
Sbjct 220 ---TINYNAAAGGTVMRQVDGTPYINQTDLNYSDAGGQPRAGGVTGTRYNIDNSSQLSGT 276
Query 292 GKSISFRNIAAHSHIQRWLDLALAGGSRYSDYINSQFDV----SRLKHTTSPLYLGSDRQ 347
++ ++ +Q WL+ GG+RY + I + F V +RL+ P YLG +
Sbjct 277 AEAADINSLRRAFRLQEWLERNARGGTRYIESILAHFGVKSSDARLQR---PEYLGGSK- 332
Query 348 YLGSNVIYQTTGAGDSSSPLGAFAGQA---SGGETFRQRSYHFGENGYFVVMASLVPDVI 404
G VI + +++ P+G AG SGG F+ Y+ E+G+ + + S+ P+
Sbjct 333 --GKMVISEVLSTAETTLPVGNMAGHGISVSGGNEFK---YNVEEHGWIIGIISVTPETA 387
Query 405 YSRGMDPFNREKTLGDVYVPALDNIAMEPLMVEQVDALPSLLSLQRGTDSVYTLSFRPDK 464
Y +G+ + D + P NI + ++ +++ A +
Sbjct 388 YQQGIHRSLSKLDRLDYFWPTFANIGEQEVLGKEIYA---------------------NG 426
Query 465 LIKNSALGYVPAWSKVMQSTSRAHGRLTTDLKYWLLNRDY 504
+ GYVP +++ SR G + T L YW L R +
Sbjct 427 INIGETFGYVPRYAEYKFLNSRVAGEMRTSLDYWHLGRKF 466
> Pichovirinae_66_Microbialites_001_Microviridae_AG0132_putative.VP1
Length=518
Score = 79.0 bits (193), Expect = 2e-17, Method: Compositional matrix adjust.
Identities = 106/522 (20%), Positives = 202/522 (39%), Gaps = 100/522 (19%)
Query 9 SRNKKSNVNLSHVSPTTLAPGNLIPISFTRIFAGDDLRFKPSAFVQAMPMNAPLVNGFKL 68
S+ +N +LSH ++ G L+P + GD +P ++ +P+ AP+++ ++
Sbjct 15 SKVDSNNFDLSHDVKSSFKMGQLVPTNVIECMPGDLFTIRPQNMLRFLPLIAPVMHRVEV 74
Query 69 CLEYFFVPDRLY--NWELLMDNTGVTDDPDKVKFPQISSPAEYSTGTIKFSLNSQSAARG 126
YFFVP+RL +WE + TG +D + +P F+ Q
Sbjct 75 TTHYFFVPNRLLWADWEKWI--TGESD---------VEAPY--------FNFIDQG---- 111
Query 127 DAARLANSIVQPGSLADYCGFPVGLFPTYDIVSDTDDRNQFCALKLLGVLDIFYHYYVNQ 186
++ G+L D+ G+P+ + SD + + L I+ YY +Q
Sbjct 112 ---------IEVGTLGDHLGYPLS-----SVSSDV----KASPMPLAAYQKIWDEYYRDQ 153
Query 187 QIERFPTASFTPTLNSGSEDERNVD-------YPVTMLRSFLDFVKRSPNP----ASAIG 235
++ FTP + + + ++ + S L F ++ + +SA
Sbjct 154 NLQ---DERFTPLVPGDNNSQLALNQQCYRRAWMHDYFTSCLPFAQKGDSVQLPISSAEN 210
Query 236 EWATSNPN----GNPIFGTWSWFCSRASIFQRCLPPYYLESWLATSGYEDSEIKVD---- 287
+ +P G + + ++ PP Y+++ + +D+ ++ +
Sbjct 211 QIVQLDPTATAPGTFVLADGQASATPGNVHIINGPPPYVQTVVDD---DDNPLQFEPRGS 267
Query 288 --LDADGKSISFRNIAAHSHIQRWLDLALAGGSRYSDYINSQFDV-SRLKHTTSPLYLGS 344
+D ++ + +Q WL+ G+RY + I S F V S P YLG
Sbjct 268 LVVDVQADAVDINTVRRAFRLQEWLEKNARAGTRYIESILSHFGVRSSDARLQRPEYLGG 327
Query 345 DRQYLGSNVIYQTTGAGDSSSPLGAFAGQ---ASGGETFRQRSYHFGENGYFVVMASLVP 401
+ + + + T D++ +G AG A GG T + Y E+G+ + + ++ P
Sbjct 328 TKGNMVISEVLATAETTDANVAVGTMAGHGITAHGGNTIK---YRCEEHGWIIGIINVQP 384
Query 402 DVIYSRGM-DPFNREKTLGDVYVPALDNIAMEPLMVEQVDALPSLLSLQRGTDSVYTLSF 460
Y +G+ F+R L D P NI G VY
Sbjct 385 VTAYQQGLPREFSRFDRL-DYPWPVFANI---------------------GEQEVYNKEL 422
Query 461 RPDKLIKNSALGYVPAWSKVMQSTSRAHGRLTTDLKYWLLNR 502
+ + GY+P ++++ SR G + L +W L R
Sbjct 423 YANSPAPDEIFGYIPRYAEMKFKNSRVAGEMRDTLDFWHLGR 464
> Pichovirinae_JCVI_003_Microviridae_AG0344_putative.VP1
Length=505
Score = 78.2 bits (191), Expect = 5e-17, Method: Compositional matrix adjust.
Identities = 111/510 (22%), Positives = 182/510 (36%), Gaps = 88/510 (17%)
Query 9 SRNKKSNVNLSHVSPTTLAPGNLIPISFTRIFAGDDLRFKPSAFVQAMPMNAPLVNGFKL 68
+ + + +L+H + G L+PI GD++ + V+ P+ AP+++ F +
Sbjct 13 KKERYNTFDLTHDVKMSGRMGELLPIMNLECIPGDNITLGADSMVKFAPLLAPVMHRFNV 72
Query 69 CLEYFFVPDRLY--NWELLMDNTGVTDDPDKVKFPQISSPAEYSTGTIKFSLNSQSAARG 126
+ YFFVP+R+ WE M TD FP I + ++ +F
Sbjct 73 TMHYFFVPNRIIWDGWEDFM-----TDVDTPPAFPIIEVNSGWTAAQQRF---------- 117
Query 127 DAARLANSIVQPGSLADYCGFP--VGLFPTYDIVSDTDDRNQFCALKLLGVLDIFYHYYV 184
ADY G P PT AL ++ +Y
Sbjct 118 ---------------ADYLGIPPNTAGVPT-----------NVSALPFAAYQCVYNEWYR 151
Query 185 NQQIERFPTASFTPTLNSGSEDERNVDYPVTMLRSFLDFVKRSPNPASAIGEWATSNPNG 244
+Q + A L +G N+ V LR+F S P + G A P G
Sbjct 152 DQNL----VAEVPYQLINGPNPRTNL--AVMRLRAFEHDYFTSALPFAQKGN-AVDIPLG 204
Query 245 NPIFGTWSWFCSRASIFQRCLPPYYLESWLATSGYEDSEIKVDLDADG-----------K 293
+ + + + Y S TS EI V D D
Sbjct 205 DITLNSDWYLDGSVPKLENSTGGYV--SGNVTSDNATGEINVGSDNDPFAYDPDGSLEVT 262
Query 294 SISFRNIAAHSHIQRWLDLALAGGSRYSDYINSQFDV-SRLKHTTSPLYLGSDRQYLGSN 352
S + ++ +Q WL+L GGSRY + I + FDV S + P Y+ +Q + +
Sbjct 263 STTITDLRRAFKLQEWLELLARGGSRYIEQIKTFFDVKSSDQRLQRPEYITGTKQPVIIS 322
Query 353 VIYQTTGAGDSSSPLGAFAGQASGGETFRQRSYHFGENGYFVVMASLVPDVIYSRGMDPF 412
I TTG + P G +G T Y+ E+G+ + + S+ P+ Y +G+
Sbjct 323 EILNTTGE-TAGLPQGNMSGHGVAVGTGNVGKYYCEEHGFIIGILSVTPNTAYQQGIPKH 381
Query 413 NREKTLGDVYVPALDNIAMEPLMVEQVDALPSLLSLQRGTDSVYTLSFRPDKLIKNSALG 472
D Y P +I +P++ ++ A + P + G
Sbjct 382 YLRTDKYDFYWPQFAHIGEQPIVNNEIYA------------------YDP---TGDETFG 420
Query 473 YVPAWSKVMQSTSRAHGRLTTDLKYWLLNR 502
Y P +++ SR G + L W R
Sbjct 421 YTPRYAEYKYMPSRVAGEFRSSLDEWHAAR 450
> Alpavirinae_Human_gut_31_037_Microviridae_AG0297_putative.VP1
Length=668
Score = 74.7 bits (182), Expect = 8e-16, Method: Compositional matrix adjust.
Identities = 76/310 (25%), Positives = 131/310 (42%), Gaps = 70/310 (23%)
Query 291 DGK-SISFRNIA--AHSHIQRWLDLALAGGSRYSDYINSQFDVSRLKHTTSPLYLGSDRQ 347
DGK +I N+A ++++ R +A++ GS Y ++ + + S + T +P+Y G
Sbjct 413 DGKFTIDALNLANKVYNYLNR---IAISDGS-YRSWLETTWTGSYTERTETPIYCGGSSA 468
Query 348 YLGSNVIYQTTGAGDSSSPLGAFAGQASGGETFRQRSY---HFGENGYFVVMASLVPDVI 404
+ + T+ A D PLG+ AG+ G ++ + Y E GY + + S+ P +
Sbjct 469 EIIFQEVVSTSAATDE--PLGSLAGR--GTDSNHKGGYVTVRATEPGYLIGITSITPRLD 524
Query 405 YSRGMDPFNREKTLGDVYVPALDNIAMEPLMVEQVDALPSLLSLQRGTDSVYTLSFRPDK 464
Y++G ++ D++ PALD I + L E + A GT + T+ +
Sbjct 525 YTQGNQWDVNLDSIDDLHKPALDGIGFQDLSAELLHA---------GTTQINTI----ND 571
Query 465 LIKNSALGYVPAWSKVMQSTSRAHGRLTTDLKYWLLNRDYGVDIDQIKNTGVIGVDVLKN 524
I +G PAW M + +RA+G T+ + +L+R Y +D KNT
Sbjct 572 TITQKFIGKQPAWIDYMTNINRAYGNFRTNENFMILSRQYSLDYK--KNT---------- 619
Query 525 LASELQQAYQAGYISFEQMDALVTLFSRIQALPEYTPYVLPNAYNDVFADTSNQAQNFVL 584
+ + T Y+ P+ YN +FAD S AQNF +
Sbjct 620 -------------------------------IKDMTTYIDPDLYNGIFADQSFDAQNFWV 648
Query 585 TCTFSMSCNR 594
+ R
Sbjct 649 QIGIEIEARR 658
Score = 63.2 bits (152), Expect = 3e-12, Method: Compositional matrix adjust.
Identities = 54/187 (29%), Positives = 85/187 (45%), Gaps = 24/187 (13%)
Query 13 KSNVNLSHVSPTTLAPGNLIPISFTRIFAGDDLRFKPSAFVQAMPMNAPLVNGFKLCLEY 72
+S NLS V T + G L+P + D + + + V P PL FKL +
Sbjct 26 RSTHNLSTVFRNTQSVGTLVPFISIPMCKDDTFKIRLTPNVLTHPTVGPLFGSFKLQNDI 85
Query 73 FFVPDRLYNWELLMDNTGVTDDPDKVKFPQISSPA--EYSTGTIKFSLNSQSAARGDAAR 130
FF P RLYN L + + + ++K P+++ A +Y+T K
Sbjct 86 FFCPFRLYNSWLHNNKNKIGLNMSQIKLPKVTLTAYKQYATDVKK--------------- 130
Query 131 LANSIVQPGSLADYCGFP-VGLFPTYDIVSDTDDRNQFCALKLLGVLDIFYHYYVNQQIE 189
++ +QP SL Y G VG+ D + +RN F A+ +L DIF +YY N+Q +
Sbjct 131 --DNPLQPSSLLTYLGISNVGMINGSDPFA---ERN-FNAVPMLAYYDIFKNYYANKQED 184
Query 190 RFPTASF 196
+F +F
Sbjct 185 KFYVRNF 191
> Gokush_gi|17402851|ref|NP_510872.1|_hypothetical_protein_PhiCPG1p2_[Guinea_pig_Chlamydia_phage]
Length=553
Score = 73.2 bits (178), Expect = 2e-15, Method: Compositional matrix adjust.
Identities = 119/536 (22%), Positives = 204/536 (38%), Gaps = 98/536 (18%)
Query 9 SRNKKSNVNLSHVSPTTLAPGNLIPISFTRIFAGDDLRFKPSAFVQAMPMNAPLVNGFKL 68
+R ++S+ + S TT G LIPI + GD K + + PL++ +L
Sbjct 21 ARIQRSSFDRSCGLKTTFDAGYLIPIFCDEVLPGDTFSLKEAFLARMATPIFPLMDNLRL 80
Query 69 CLEYFFVPDRLYNWELLMDNTGVTDDP-DKVKF--PQISSPAEYSTGTIKFSLNSQSAAR 125
+YFFVP RL W G D+P D F P +++P S G I+
Sbjct 81 DTQYFFVPLRLL-WSNFQKFCGEQDNPGDSTDFLTPVLTAP---SGGFIE---------- 126
Query 126 GDAARLANSIVQPGSLADYCGFPVGLFPTYDIVSDTDDRNQFCALKLLGVLDIFYHYYVN 185
GS+ DY G P + + A I+ YY +
Sbjct 127 -------------GSIHDYLGLPTKVAGI-----------ECVAFWHRAYNLIWNQYYRD 162
Query 186 QQIERFPTASFTPT----LNSGSEDERNVDYPVTMLRSFLDFVKRSPNPASAIG------ 235
+ I+ T +N+ +R Y S L + ++ P +G
Sbjct 163 ENIQESVDVEMGDTTSNEVNNYKLLKRGKRY--DYFTSCLPWPQKGPAVTIGVGGIVPVQ 220
Query 236 ----EWATSNPNGNPIFGTWSWFCSRASIFQRCLPP--------------YYLESWLATS 277
+W S+ NPI + SW S F P YY++ +
Sbjct 221 GLGIQWGNSSAP-NPITAS-SWINSVNPTFINSTTPTPTGTNKILNYGQAYYIKKPGEAT 278
Query 278 GYEDSEIKVDLDADGKSISFRNIAAHSHIQRWLDLALAGGSRYSDYINSQFDV----SRL 333
VDL + ++ ++ +Q+ + GG+RY + I S F+V +RL
Sbjct 279 TDPTPRAYVDLGST-SPVTINSLREAFQLQKLYERDARGGTRYIEIIRSHFNVQSPDARL 337
Query 334 KHTTSPLYLGSDRQYLGSNVIYQTTGAGDSSSPLGAFAGQASGGETFRQRSYHFGENGYF 393
+ YLG + + I QT+ DS+SP G A + + R + F E+G
Sbjct 338 QRAE---YLGGSSTPVNISPIPQTSST-DSTSPQGNLAAYGTAIGSKRVFTKSFTEHGVI 393
Query 394 VVMASLVPDVIYSRGMDPFNREKTLGDVYVPALDNIAMEPLMVEQVDAL-PSLLSLQRGT 452
+ +AS+ D+ Y +G+D +T D Y PAL ++ + ++ +++ P++ Q G
Sbjct 394 LGLASVRADLNYQQGLDRMWSRRTRWDFYWPALSHLGEQAVLNKEIYCQGPAVKDAQNG- 452
Query 453 DSVYTLSFRPDKLIKNSALGYVPAWSKVMQSTSRAHGRL----TTDLKYWLLNRDY 504
+ ++ GY +++ TS+ G+ T L W L + +
Sbjct 453 ----------NVVVDEQVFGYQERFAEYRYKTSKITGKFRSNATGSLDAWHLAQQF 498
Lambda K H a alpha
0.319 0.135 0.406 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 55697121