bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: Microviridae_proteins.fasta
575 sequences; 147,441 total letters
Query= Contig-37_CDS_annotation_glimmer3.pl_2_2
Length=731
Score E
Sequences producing significant alignments: (Bits) Value
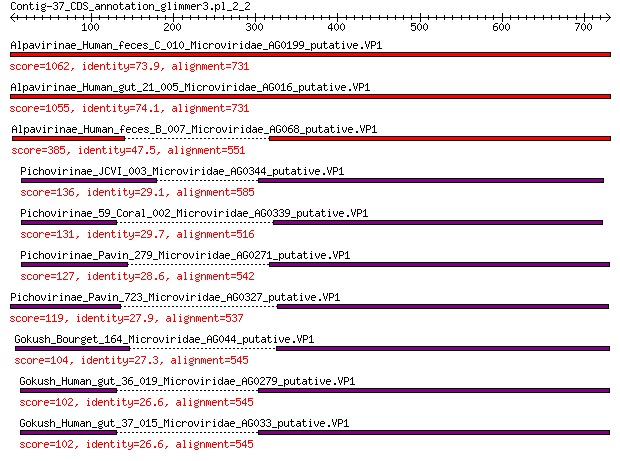
Alpavirinae_Human_feces_C_010_Microviridae_AG0199_putative.VP1 1062 0.0
Alpavirinae_Human_gut_21_005_Microviridae_AG016_putative.VP1 1055 0.0
Alpavirinae_Human_feces_B_007_Microviridae_AG068_putative.VP1 385 3e-123
Pichovirinae_JCVI_003_Microviridae_AG0344_putative.VP1 136 7e-36
Pichovirinae_59_Coral_002_Microviridae_AG0339_putative.VP1 131 5e-34
Pichovirinae_Pavin_279_Microviridae_AG0271_putative.VP1 127 5e-33
Pichovirinae_Pavin_723_Microviridae_AG0327_putative.VP1 119 3e-30
Gokush_Bourget_164_Microviridae_AG044_putative.VP1 104 3e-25
Gokush_Human_gut_36_019_Microviridae_AG0279_putative.VP1 102 1e-24
Gokush_Human_gut_37_015_Microviridae_AG033_putative.VP1 102 1e-24
> Alpavirinae_Human_feces_C_010_Microviridae_AG0199_putative.VP1
Length=731
Score = 1062 bits (2747), Expect = 0.0, Method: Compositional matrix adjust.
Identities = 540/737 (73%), Positives = 595/737 (81%), Gaps = 12/737 (2%)
Query 1 MAQNIFDATFDANNRIDVNSFDWSHVNNLTTDFGRITPVFCELVPAKGSLRINPEFGLEL 60
MAQNIFDATFDANNRIDVNSFDWSHVNNLTTDFGRITPVFCELVPAKGSLRINPEFGLEL
Sbjct 1 MAQNIFDATFDANNRIDVNSFDWSHVNNLTTDFGRITPVFCELVPAKGSLRINPEFGLEL 60
Query 61 MPMVFPVQTRMFARLNFFKVTLRSMWEDYSDFISNFRDDLEEPYILPGSQNFTTMLGTGS 120
MPMVFPVQTRMFARLNFFKVTLRSMWEDYSDFISNFRDDLEEPYILP S F M TGS
Sbjct 61 MPMVFPVQTRMFARLNFFKVTLRSMWEDYSDFISNFRDDLEEPYILPDSFRFEKMCKTGS 120
Query 121 LGDYLGVPTRNAGAEVSLMAHPSQCAANSMLVFSPSVTLQEVCSSFLSNTIPKANCA--L 178
LGDYLG+PT + + + CAA S V S S + + S S+ IP + CA L
Sbjct 121 LGDYLGLPTFGTKSSRVYNFNDNPCAARSRTV-SKSDIGKVMASVRSSSGIPDSVCASTL 179
Query 179 SFGNSDGRKQVAYFSITLQGYNNAMRSYKLTVGIKKANDKDR-NSYSGAIVFSSATNVYD 237
+ G S K V++ S+ + D + +G V S N Y
Sbjct 180 TPGVS---KTVSFTSVNSSNVVPSDIIQIGIDITISGGDSAAFDGTTGYFVLLSKDNKYL 236
Query 238 RFENVTFVWDES-TSSYVTTISFSDKQYSAVSVLLQTEPGIASSSLWQFGLNTASAV--A 294
+T ++ ++++++ S S + L+ P ++ + F S V
Sbjct 237 FDIPITLKAEKGVVMAHISSVEASKFGSSYGTPLVIATPD--TTDVISFDNIKFSVVYTT 294
Query 295 FVEESETTYETTPFATQNKPANLRLLAYRFRAYESVYNAYYRDIRNNPFIVDGRPVYNKW 354
+ +S+ +Y T PFAT+++P RLLAYRFRAYESVYNAYYRDIRNNPF+V+GRPVYNKW
Sbjct 295 YSLQSDVSYLTYPFATESRPDISRLLAYRFRAYESVYNAYYRDIRNNPFVVNGRPVYNKW 354
Query 355 LPSMKGGADNTTYSLHQCNWERDFLTTAVPNPQQGANAPLVGLTVGDVVTRADDGTYSIQ 414
LP+MKGGAD T Y L QCNWERDFLTTAVPNPQQGANAPLVGL +GDVVTR++DGTYS+Q
Sbjct 355 LPTMKGGADTTLYELRQCNWERDFLTTAVPNPQQGANAPLVGLVMGDVVTRSEDGTYSVQ 414
Query 415 KQTVLVDEDGSKYGISYKVSEDGESLVGVDYDPVSEKTPVTAINSYAELAALTTTEGSGF 474
KQTVLVDEDGSKYG+SYKVSEDGE LVGVDYDPVSEKTPVTAI SYAELAAL T +GSGF
Sbjct 415 KQTVLVDEDGSKYGVSYKVSEDGERLVGVDYDPVSEKTPVTAIGSYAELAALATEQGSGF 474
Query 475 TIETLRYVNAYQKFLELNMRKGFSYKQIMQGRWDIDIRFDELLMPEFIGGISRELSMRTV 534
TIETLRYVNAYQKFLELNMRKGFSYKQIMQGRWDIDIRFDELLMPEFIGGISRELSMRTV
Sbjct 475 TIETLRYVNAYQKFLELNMRKGFSYKQIMQGRWDIDIRFDELLMPEFIGGISRELSMRTV 534
Query 535 EQTVDQQSTSSQGQYAEALGSKSGIAGVYGSTSNNIEVFCDEESYIIGLLTVTPVPIYTQ 594
EQTVDQQS SS+GQYAEALGSK+GIAGVYGSTSNNIEVFCDEESYIIGLLTVTPVPIYTQ
Sbjct 535 EQTVDQQSGSSRGQYAEALGSKTGIAGVYGSTSNNIEVFCDEESYIIGLLTVTPVPIYTQ 594
Query 595 MLSKDFLYNGLLDHYQPEFDRIGFQPITYKEVCPLNLGVADTVNKANQTFGYQRPWYEYV 654
+L KDF YNGLLDHYQPEFDRIGFQPITYKE+CP+N+ D+ N+ N+TFGYQRPWYEYV
Sbjct 595 LLPKDFTYNGLLDHYQPEFDRIGFQPITYKEICPMNIVAGDSANQLNKTFGYQRPWYEYV 654
Query 655 AKYDSAHGLFRTNMKNFVMSRVFSGLPQLGQQFLLVDPDTVNQVFSVTEYTDKIFGYVKF 714
AKYDSAHGLFRTNMKNF+MSRVFSGLPQLGQQFLLVDPDTVNQVFSVTEYTDKIFGYVKF
Sbjct 655 AKYDSAHGLFRTNMKNFIMSRVFSGLPQLGQQFLLVDPDTVNQVFSVTEYTDKIFGYVKF 714
Query 715 NATARLPISRVAIPRLD 731
NATARLPISRVAIPRLD
Sbjct 715 NATARLPISRVAIPRLD 731
> Alpavirinae_Human_gut_21_005_Microviridae_AG016_putative.VP1
Length=742
Score = 1055 bits (2727), Expect = 0.0, Method: Compositional matrix adjust.
Identities = 542/752 (72%), Positives = 593/752 (79%), Gaps = 31/752 (4%)
Query 1 MAQNIFDATFDANNRIDVNSFDWSHVNNLTTDFGRITPVFCELVPAKGSLRINPEFGLEL 60
MAQNIFDATFDANNRIDVNSFDWSHVNNLTTDFGRITPVFCELVPAKGSLRINPEFGLEL
Sbjct 1 MAQNIFDATFDANNRIDVNSFDWSHVNNLTTDFGRITPVFCELVPAKGSLRINPEFGLEL 60
Query 61 MPMVFPVQTRMFARLNFFKVTLRSMWEDYSDFISNFRDDLEEPYILPGSQNFTTMLGTGS 120
MPMVFPVQTRMFARLNFFKVTLRSMWEDYSDFISNFRDDLEEPYI +F M TGS
Sbjct 61 MPMVFPVQTRMFARLNFFKVTLRSMWEDYSDFISNFRDDLEEPYINCSEISFPKMFTTGS 120
Query 121 LGDYLGVPTRNAGAEVS---LMAHPSQCAANSMLVFSPSVTLQEVCSSFLSN----TIPK 173
LGDYLGVPTR + S + + ++ + + SPS + + S+ N TIP
Sbjct 121 LGDYLGVPTRKSYHSSSTYISRSMSACSSSKKIALVSPSYSWDTIISNLSFNSAISTIPG 180
Query 174 ANCALSFGNSDGRKQVAYFSITLQGYNNAMRSYKLTVGIKKANDKDRNSYSGAIVFS--- 230
+CA ++ S + +Q Y NA + L ND R+ S ++
Sbjct 181 VSCADAYDKSSTVILKTSSPVKVQRYKNASVEFYL------LNDVKRDYVSHGLLVGITP 234
Query 231 SATNVYDRFENVTFVWDESTSSYVTTIS-----FSDKQYSA----VSVLLQTEPGIASSS 281
T+++ N + + V T+ FSD+ A VLL P S
Sbjct 235 GNTSIHTWEFNAPVNTKGNLTRMVVTLGNLANVFSDQAADADGIDFYVLL---PSADMPS 291
Query 282 LWQFGLNTASAVAFVEESETTYETTPFATQNKPANL--RLLAYRFRAYESVYNAYYRDIR 339
L L+T + EESE T++ PF +Q RLLAYRFRAYE+VYNAYYRDIR
Sbjct 292 LTDIALSTLTFFESPEESEITFDDFPFKSQEAGTKSYPRLLAYRFRAYEAVYNAYYRDIR 351
Query 340 NNPFIVDGRPVYNKWLPSMKGGADNTTYSLHQCNWERDFLTTAVPNPQQGANAPLVGLTV 399
NNPF+V+GRPVYNKWLP+MKGGAD T Y LHQCNWERDFLTTAVPNPQQGANAPLVGLTV
Sbjct 352 NNPFVVNGRPVYNKWLPTMKGGADETMYQLHQCNWERDFLTTAVPNPQQGANAPLVGLTV 411
Query 400 GDVVTRADDGTYSIQKQTVLVDEDGSKYGISYKVSEDGESLVGVDYDPVSEKTPVTAINS 459
GDVVTR++DGT S+QKQTVLVDEDGSKYGISYKVSEDGE LVGVDYDPVSEKTPVTAINS
Sbjct 412 GDVVTRSEDGTLSVQKQTVLVDEDGSKYGISYKVSEDGERLVGVDYDPVSEKTPVTAINS 471
Query 460 YAELAALTTTEGSGFTIETLRYVNAYQKFLELNMRKGFSYKQIMQGRWDIDIRFDELLMP 519
YAELAAL T +GSGFTIETLRYVNAYQKFLELNMRKGFSYKQIMQGRWDIDIRFDELLMP
Sbjct 472 YAELAALATEQGSGFTIETLRYVNAYQKFLELNMRKGFSYKQIMQGRWDIDIRFDELLMP 531
Query 520 EFIGGISRELSMRTVEQTVDQQSTSSQGQYAEALGSKSGIAGVYGSTSNNIEVFCDEESY 579
EFIGGISRELSMRTVEQTVDQQS SSQGQYAEALGSK+GIAGVYGSTSNNIEVFCDEESY
Sbjct 532 EFIGGISRELSMRTVEQTVDQQSASSQGQYAEALGSKTGIAGVYGSTSNNIEVFCDEESY 591
Query 580 IIGLLTVTPVPIYTQMLSKDFLYNGLLDHYQPEFDRIGFQPITYKEVCPLNLGVADTVNK 639
IIGLLTVTPVPIY+Q+L KDF YNGLLDHYQPEFDRIGFQPITYKEVCPLN +++ +N
Sbjct 592 IIGLLTVTPVPIYSQLLPKDFTYNGLLDHYQPEFDRIGFQPITYKEVCPLNFDISN-MND 650
Query 640 ANQTFGYQRPWYEYVAKYDSAHGLFRTNMKNFVMSRVFSGLPQLGQQFLLVDPDTVNQVF 699
N+TFGYQRPWYEYVAKYD+AHGLFRT++K FVM R FSGLPQLGQQFLLVDPDTVNQVF
Sbjct 651 MNKTFGYQRPWYEYVAKYDNAHGLFRTDLKKFVMHRTFSGLPQLGQQFLLVDPDTVNQVF 710
Query 700 SVTEYTDKIFGYVKFNATARLPISRVAIPRLD 731
SVTEYTDKIFGYVKFNATARLPISRVAIPRLD
Sbjct 711 SVTEYTDKIFGYVKFNATARLPISRVAIPRLD 742
> Alpavirinae_Human_feces_B_007_Microviridae_AG068_putative.VP1
Length=780
Score = 385 bits (988), Expect = 3e-123, Method: Compositional matrix adjust.
Identities = 190/416 (46%), Positives = 279/416 (67%), Gaps = 16/416 (4%)
Query 317 LRLLAYRFRAYESVYNAYYRDIRNNPFIVDGRPVYNKWLPSMKGGADNTT-YSLHQCNWE 375
+++ AY FRAY+++YNAY R+ RNNPFI++G+ YN+W+ + +GG D+ T L NW+
Sbjct 380 IKVSAYPFRAYQAIYNAYIRNTRNNPFILNGKKTYNRWITTDEGGEDSLTPLDLLYANWQ 439
Query 376 RDFLTTAVPNPQQGANAPLVGLTVGDVVTRADDGTYSIQKQTVLVDEDGSKYGISYKVSE 435
D TTA+ PQQG APLVGLT + V+ D G T +VDEDG+ Y + ++
Sbjct 440 SDAYTTALTAPQQGV-APLVGLTTYESVSLNDAGHTVTTVNTAIVDEDGNAYKVDFE--S 496
Query 436 DGESLVGVDYDPVSEKTPVTAINSYAELAALTTTEGSGFTIETLRYVNAYQKFLELNMRK 495
+GE+L GV+Y P+ AIN + +L + SG +I R VNAYQ++LELN +
Sbjct 497 NGEALKGVNYTPLKAGE---AIN----MQSLVSPVTSGISINDFRNVNAYQRYLELNQFR 549
Query 496 GFSYKQIMQGRWDIDIRFDELLMPEFIGGISRELSMRTVEQTVDQQSTSSQGQYAEALGS 555
GFSYK+I++GR+D+++R+D L MPE++GGI+R++ + + QTV+ T+ G Y +LGS
Sbjct 550 GFSYKEIIEGRFDVNVRYDALNMPEYLGGITRDIVVNPITQTVE---TTDTGSYVGSLGS 606
Query 556 KSGIAGVYGSTSNNIEVFCDEESYIIGLLTVTPVPIYTQMLSKDFLYNGLLDHYQPEFDR 615
++G+A +G++ +I VFCDEES ++G++ V P+P+Y +L K Y LD + PEFD
Sbjct 607 QAGLATCFGNSDGSISVFCDEESIVMGIMYVMPMPVYDSILPKWLTYRERLDSFNPEFDH 666
Query 616 IGFQPITYKEVCPLNLGVADTVNKANQTFGYQRPWYEYVAKYDSAHGLFRTNMKNFVMSR 675
IG+QPI KE+ + ++ N FGYQRPWYEYV K D AHGLF ++++NF+M R
Sbjct 667 IGYQPIYLKELAAIQ--AFESGKDLNTVFGYQRPWYEYVQKVDRAHGLFLSSLRNFIMFR 724
Query 676 VFSGLPQLGQQFLLVDPDTVNQVFSVTEYTDKIFGYVKFNATARLPISRVAIPRLD 731
F P+LG+ F ++ P +VN VFSVTE +DKI G + F+ TA+LPISRV +PRL+
Sbjct 725 SFENAPELGKSFTVMQPGSVNNVFSVTEVSDKILGQIHFDCTAQLPISRVVVPRLE 780
Score = 143 bits (360), Expect = 3e-37, Method: Compositional matrix adjust.
Identities = 72/141 (51%), Positives = 92/141 (65%), Gaps = 7/141 (5%)
Query 4 NIFDATFDANNRIDVNSFDWSHVNNLTTDFGRITPVFCELVPAKGSLRINPEFGLELMPM 63
++F+ D N + NSFDWSH NN TTD GRITPVF ELVP S+RI PEFGL MPM
Sbjct 3 SVFNKIGDVKNDVKRNSFDWSHDNNFTTDLGRITPVFTELVPPNSSVRIKPEFGLRFMPM 62
Query 64 VFPVQTRMFARLNFFKVTLRSMWEDYSDFISNFRDDLEE---PYILPGSQNFTT--MLGT 118
+FP+QT+M A L+FFKV LR++W+DY DFIS D+ EE P+I ++F+ L
Sbjct 63 MFPIQTKMKAYLSFFKVPLRTLWKDYMDFIS--ADNTEEYQPPFIKFSGKSFSEGGALSE 120
Query 119 GSLGDYLGVPTRNAGAEVSLM 139
LGDY+G+P N +M
Sbjct 121 SGLGDYIGMPVSNIDTNWQVM 141
> Pichovirinae_JCVI_003_Microviridae_AG0344_putative.VP1
Length=505
Score = 136 bits (343), Expect = 7e-36, Method: Compositional matrix adjust.
Identities = 128/431 (30%), Positives = 198/431 (46%), Gaps = 62/431 (14%)
Query 303 YETTPFATQNKPANLRLLAYRFRAYESVYNAYYRD---IRNNPFIVDGRPVYNKWLPSMK 359
Y P T P N+ L F AY+ VYN +YRD + P+ + P L M+
Sbjct 120 YLGIPPNTAGVPTNVSALP--FAAYQCVYNEWYRDQNLVAEVPYQLINGPNPRTNLAVMR 177
Query 360 GGADNTTYSLHQCNWERDFLTTAVPNPQQGANAPLVGLTVGDVVTRAD---DGTYSIQKQ 416
A +E D+ T+A+P Q+G NA V + +GD+ +D DG S+ K
Sbjct 178 LRA-----------FEHDYFTSALPFAQKG-NA--VDIPLGDITLNSDWYLDG--SVPKL 221
Query 417 TVLVDEDGSKYGISYKVSED---GESLVGVDYDPVSEKTPVTAINSYAELAALTTTEGSG 473
E+ + +S V+ D GE VG D DP + Y +L T
Sbjct 222 -----ENSTGGYVSGNVTSDNATGEINVGSDNDPFA----------YDPDGSLEVTST-- 264
Query 474 FTIETLRYVNAYQKFLELNMRKGFSYKQIMQGRWDIDIRFDELLMPEFIGGISRELSMRT 533
TI LR Q++LEL R G Y + ++ +D+ L PE+I G + + +
Sbjct 265 -TITDLRRAFKLQEWLELLARGGSRYIEQIKTFFDVKSSDQRLQRPEYITGTKQPVIISE 323
Query 534 VEQTVDQQSTSSQGQYAEALGSKSGIAGVYGSTSNNIEVFCDEESYIIGLLTVTPVPIYT 593
+ T + + QG + SG GV T N + +C+E +IIG+L+VTP Y
Sbjct 324 ILNTTGETAGLPQG-------NMSG-HGVAVGTGNVGKYYCEEHGFIIGILSVTPNTAYQ 375
Query 594 QMLSKDFLYNGLLDHYQPEFDRIGFQPITYKEVCPLNLGVADTVNKANQTFGYQRPWYEY 653
Q + K +L D Y P+F IG QPI E+ + ++TFGY + EY
Sbjct 376 QGIPKHYLRTDKYDFYWPQFAHIGEQPIVNNEIYAYD-------PTGDETFGYTPRYAEY 428
Query 654 VAKYDSAHGLFRTNMKNFVMSRVFSGLPQLGQQFLLVDPDTVNQVFSVTEYTDKIFGYV- 712
G FR+++ + +R+FS LP L Q F+ VD D +++F+VT+ D ++ +V
Sbjct 429 KYMPSRVAGEFRSSLDEWHAARIFSSLPTLSQDFIEVDADDFDRIFAVTDGDDNLYMHVL 488
Query 713 -KFNATARLPI 722
K A ++P+
Sbjct 489 NKVKARRQMPV 499
Score = 54.7 bits (130), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 42/175 (24%), Positives = 78/175 (45%), Gaps = 17/175 (10%)
Query 14 NRIDV-----NSFDWSHVNNLTTDFGRITPVF-CELVPAKGSLRINPEFGLELMPMVFPV 67
NR+ V N+FD +H ++ G + P+ E +P ++ + + ++ P++ PV
Sbjct 8 NRVSVKKERYNTFDLTHDVKMSGRMGELLPIMNLECIPGD-NITLGADSMVKFAPLLAPV 66
Query 68 QTRMFARLNFFKVTLRSMWEDYSDFISNFRDDLEEPYILPGSQ-NFTTMLGTGSLGDYLG 126
R +++F V R +W+ + DF++ D++ P P + N DYLG
Sbjct 67 MHRFNVTMHYFFVPNRIIWDGWEDFMT----DVDTPPAFPIIEVNSGWTAAQQRFADYLG 122
Query 127 VPTRNAGAEVSLMAHP---SQCAANSMLVFSPSVTLQEVCSSFLSNTIPKANCAL 178
+P AG ++ A P QC N + + EV ++ P+ N A+
Sbjct 123 IPPNTAGVPTNVSALPFAAYQCVYNEW--YRDQNLVAEVPYQLINGPNPRTNLAV 175
> Pichovirinae_59_Coral_002_Microviridae_AG0339_putative.VP1
Length=522
Score = 131 bits (329), Expect = 5e-34, Method: Compositional matrix adjust.
Identities = 115/414 (28%), Positives = 188/414 (45%), Gaps = 46/414 (11%)
Query 321 AYRFRAYESVYNAYYRDIRNNPFIVDGRPVYNKWLPSMKGGADNTTY-SLHQCNWERDFL 379
A F AY+ +Y+ YYRD ++D + + GAD ++ + W+ D+
Sbjct 135 ALPFAAYQKIYDDYYRDEN----LIDKVDI--TLSDGTQSGADAIELGTMRKRAWQHDYF 188
Query 380 TTAVPNPQQGANAPLVGLTVGDVVTRADDGTYSIQKQTVLVDE------DGSKY---GIS 430
T+A+P Q+G A + T + + T +I + + DG+ G S
Sbjct 189 TSALPWTQRGPEATIPLGTSAPITWSNNTATATIVRNNTSGNPIAGYTFDGASALYTGGS 248
Query 431 YKVSEDGESLVGVDYDPVSEKTPVTAINSYAELAALTTTEGSGFTIETLRYVNAYQKFLE 490
++ D + V +D+D + + +A+LA T + +I LR Q++LE
Sbjct 249 GQLIADLPTPVSLDFD--------NSDHLFADLAGATAS-----SINDLRRAFRLQEWLE 295
Query 491 LNMRKGFSYKQIMQGRWDIDIRFDELLMPEFIGGISRELSMRTVEQT-VDQQSTSSQGQY 549
N R G Y +I+ + + L PEF+GG + +++ V QT + + QG
Sbjct 296 RNARGGARYIEIIMAHFGVRSSDARLQRPEFLGGSATPITISEVLQTSANNTEPTPQGNM 355
Query 550 AEALGSKSGIAGVYGSTSNNIEVFCDEESYIIGLLTVTPVPIYTQMLSKDFLYNGLLDHY 609
A GV +SN + C+E YIIG+++V P Y Q + K F D+Y
Sbjct 356 AGH--------GVSVGSSNYVSYKCEEHGYIIGIMSVMPKTAYQQGIPKHFKKLDKFDYY 407
Query 610 QPEFDRIGFQPITYKEVCPLNLGVADTVNKANQTFGYQRPWYEYVAKYDSAHGLFRTNMK 669
P F IG QPI +E+ N DT +TFGY + EY + HG FR ++
Sbjct 408 WPSFANIGEQPILNEELYHQN-NATDT-----ETFGYTPRYAEYKYIPSTVHGEFRDSLD 461
Query 670 NFVMSRVFSGLPQLGQQFLLVDPDTVNQVFSVTEYTDKIFGYV--KFNATARLP 721
+ M R+F P L Q F+ + D V +VF+VT + ++ Y+ + AT +P
Sbjct 462 FWHMGRIFGSKPTLNQDFIECNADDVERVFAVTAGQEHLYVYLHNEVKATRLMP 515
Score = 55.8 bits (133), Expect = 7e-10, Method: Compositional matrix adjust.
Identities = 38/119 (32%), Positives = 54/119 (45%), Gaps = 7/119 (6%)
Query 15 RIDVNSFDWSHVNNLTTDFGRITPV-FCELVPAKGSLRINPEFGLELMPMVFPVQTRMFA 73
R N+FD SH + G + P+ E VP I P++ P+ +
Sbjct 11 RPQTNTFDLSHDRKFSGKIGELMPITVMEAVPGD-KFNIKATNLTRFAPLITPIMHQASV 69
Query 74 RLNFFKVTLRSMWEDYSDFISNFRDDLEEPYILPGSQNFTTM---LGTGSLGDYLGVPT 129
+FF V R +W ++ DFIS D L +P P + N TT G+L DYLG+PT
Sbjct 70 YCHFFFVPNRILWSNWEDFISGGEDGLADP-TFP-TINITTPTNGYAVGALADYLGLPT 126
> Pichovirinae_Pavin_279_Microviridae_AG0271_putative.VP1
Length=503
Score = 127 bits (320), Expect = 5e-33, Method: Compositional matrix adjust.
Identities = 117/424 (28%), Positives = 184/424 (43%), Gaps = 60/424 (14%)
Query 317 LRLLAYRFRAYESVYNAYYRD----IRNNPFIVDGRPVYNKWLPSMKGGADNTTYSLHQC 372
+++ A F AY+++YN YYRD N + DG N+ + T +L +
Sbjct 130 VQINALPFAAYQAIYNEYYRDENLVTAVNYNLADG----NQGI--------GTFGTLRKR 177
Query 373 NWERDFLTTAVPNPQQGANAPLVGLTVGDVVTRADDGTYSIQK-----QTVLVDEDGSKY 427
+E D+ T +P Q+GA V L +GDV +A+ +I K T + + D +
Sbjct 178 AYEHDYFTACLPFAQKGA---AVDLPLGDVTLKANWNPSAIPKFVNAAGTSINNLDDVQG 234
Query 428 GISYKVSEDGESLVGVDYDPVSEKTPVTAINSYAELAALTTTEGSGFTIETLRYVNAYQK 487
G ++ G ++P+ A N L +TT I LR Q+
Sbjct 235 GTPNRIRTSGG------------QSPI-AYNPNGSLEVGSTT------INDLRRAMRLQE 275
Query 488 FLELNMRKGFSYKQIMQGRWDIDIRFDELLMPEFIGGISRELSMRTVEQTVDQQSTSSQG 547
FLE N R G Y + ++ + + L PE+I G + ++ + TV
Sbjct 276 FLEKNARGGTRYIENIKMHFGVQSSDSRLQRPEYITGTKAPVVVQEITSTVKGIEEPQGN 335
Query 548 QYAEALGSKSGIAGVYGSTSNNIEVFCDEESYIIGLLTVTPVPIYTQMLSKDFLYNGLLD 607
+A +SG G Y +E YIIG++++ P P Y Q + K +L +LD
Sbjct 336 PTGKATSLQSGHYGNYS---------VEEHGYIIGIMSIMPKPAYQQGIPKSYLKTDVLD 386
Query 608 HYQPEFDRIGFQPITYKEVCPLNLGVADTVNKANQTFGYQRPWYEYVAKYDSAHGLFRTN 667
Y P F IG Q + +E+ G T N FGY + EY + GLFRT
Sbjct 387 FYWPSFANIGEQEVLQEEI----YGYTPT---PNAVFGYIPRYAEYKYLQNRVCGLFRTT 439
Query 668 MKNFVMSRVFSGLPQLGQQFLLVDPDTVNQVFSVTE-YTDKIFGYVKFNATARLPISRVA 726
+ + + R+F+ +P L Q F+ V P ++F+ E Y D I+ +V TAR P+
Sbjct 440 LDFWHLGRIFANIPTLSQSFIEVSPANQTRIFANQEDYDDNIYIHVLNKMTARRPMPVFG 499
Query 727 IPRL 730
P L
Sbjct 500 TPML 503
Score = 42.7 bits (99), Expect = 9e-06, Method: Compositional matrix adjust.
Identities = 38/132 (29%), Positives = 56/132 (42%), Gaps = 11/132 (8%)
Query 15 RIDVNSFDWSHVNNLTTDFGRITPV-FCELVPAKGSLRINPEFGLELMPMVFPVQTRMFA 73
R N+FD +H +T G + P E VP I+ + L P+ P+ +
Sbjct 12 RPPSNTFDLTHDVKMTGGMGNLMPCCIAECVPGD-KWNISGDTYLRFAPITAPIMHSIDV 70
Query 74 RLNFFKVTLRSMWEDYSDFISNFRDDLEEPY--ILPGSQNFTTMLGTGSLGDYLGVPTRN 131
+++F V R +W ++ FI N EP I S + LGDYL VP
Sbjct 71 AIHYFFVPNRILWSNWEKFIVN------EPTGGIPTFSWDANIPATAKKLGDYLSVPPPP 124
Query 132 AGA-EVSLMAHP 142
GA +V + A P
Sbjct 125 TGATQVQINALP 136
> Pichovirinae_Pavin_723_Microviridae_AG0327_putative.VP1
Length=508
Score = 119 bits (299), Expect = 3e-30, Method: Compositional matrix adjust.
Identities = 110/412 (27%), Positives = 178/412 (43%), Gaps = 57/412 (14%)
Query 326 AYESVYNAYYRDIRNNPFIVDGRPVYNKWLPSMKGGADNTTY-------SLHQCNWERDF 378
AY+ +YN YYRD +N VD + + G +NT +L Q WE D+
Sbjct 142 AYQCIYNEYYRD-QNLQTPVDYK---------LTDGNNNTDAGDRERLTTLRQRAWEHDY 191
Query 379 LTTAVPNPQQGANAPLVGLTVGDVVTRADDGTYSIQKQTVLVDEDGSKYGISYKVSEDGE 438
T ++P Q+GA V + +G + + V V+ + ++G + +
Sbjct 192 FTASLPFAQKGA---AVDIPIGTI------------DKDVAVNFNSLEFGATTVRYNSAD 236
Query 439 SLVGVDYDPVSEKTPVTAINSYAELAALTTTEGSGFTIETLRYVNAYQKFLELNMRKGFS 498
L+ + V A + +A +T + TI LR Q++LE N R G
Sbjct 237 GLIDI-----GGSNAVNADGTPTMIAKTSTIDIEPTTINDLRRAFKLQEWLEKNARGGTR 291
Query 499 YKQIMQGRWDIDIRFDELLMPEFIGGISRELSMRTVEQTVDQQSTSSQGQYA-EALGSKS 557
Y + + + + L PE+I G+ + + V T Q QG A + S
Sbjct 292 YIENILTHFGVRSSDKRLQRPEYITGVKSPVVVSEVLNTTGQDGGLPQGNMAGHGISVTS 351
Query 558 GIAGVYGSTSNNIEVFCDEESYIIGLLTVTPVPIYTQMLSKDFLYNGLLDHYQPEFDRIG 617
G +G Y +C+E YIIG+++V P Y Q + + FL LD++ P F IG
Sbjct 352 GKSGSY---------YCEEHGYIIGIMSVMPKTAYQQGIPRTFLKTDSLDYFWPTFANIG 402
Query 618 FQPITYKEVCPLNLGVADTVNKANQTFGYQRPWYEYVAKYDSAHGLFRTNMKNFVMSRVF 677
Q + +E+ A T N AN TFGY + EY G FRT++ + + R+F
Sbjct 403 EQEVAKQEL------YAYTAN-ANDTFGYVPRYAEYKYMPSRVAGEFRTSLNYWHLGRIF 455
Query 678 SGLPQLGQQFLLVDPDTVNQVFSVTE-YTDKIFGYVKFNATARLPISRVAIP 728
+ P L F+ DP ++F+V + TD ++ +V A P+ + P
Sbjct 456 ATEPSLNSDFIECDP--TKRIFAVEDPETDVLYCHVLNKIKAVRPMPKYGTP 505
Score = 65.9 bits (159), Expect = 4e-13, Method: Compositional matrix adjust.
Identities = 40/135 (30%), Positives = 68/135 (50%), Gaps = 6/135 (4%)
Query 1 MAQNIFDATFDANNRIDVNSFDWSHVNNLTTDFGRITPVFC-ELVPAKGSLRINPEFGLE 59
M QN+F++ N+ N FD +H L+T+ G++TP+ E VP ++ E +
Sbjct 1 MGQNLFNSI--QLNKPKKNVFDLTHDVKLSTNMGQLTPILTLECVPGD-KFDLSCESLIR 57
Query 60 LMPMVFPVQTRMFARLNFFKVTLRSMWEDYSDFISNFRDDLEEPYILPGSQNFTTMLGTG 119
PM+ PV RM +++F V R +W ++ FI+ + PY+ + ++T M
Sbjct 58 FAPMIAPVMHRMDVTMHYFFVPNRILWSNWEKFITEHNSEHVAPYMAYTNGDYTAM--QK 115
Query 120 SLGDYLGVPTRNAGA 134
DY+G+P AG
Sbjct 116 KFMDYIGIPPVPAGG 130
> Gokush_Bourget_164_Microviridae_AG044_putative.VP1
Length=540
Score = 104 bits (259), Expect = 3e-25, Method: Compositional matrix adjust.
Identities = 106/414 (26%), Positives = 173/414 (42%), Gaps = 37/414 (9%)
Query 325 RAYESVYNAYYRD--IRNNPFIVDGRPVYNKWLPSMKGGADNTTYSLHQCNWERDFLTTA 382
RAY +YN ++RD ++N+ + G + T Y++ + D+ T+A
Sbjct 152 RAYNLIYNQWFRDENLQNSSVVDKG---------DGPDASPATNYTILRRGKRHDYFTSA 202
Query 383 VPNPQQGANAPLVGLTVGDVVTRADDGTYSIQKQTVLVDEDGSKYGISYKVSEDGESLVG 442
+P PQ+G A V + +G +G + T K G S +++++G++ G
Sbjct 203 LPWPQKGGTA--VTIPIGTSAPIKSNGGANFYAWTGSSGTSQLKVGTSGQLTQNGQANPG 260
Query 443 VDYDPVSEKTPVTAINSYAELAALTTTEGSGFTIETLRYVNAYQKFLELNMRKGFSYKQI 502
D + T YA+L+ T TI LR QK LE + R G Y +I
Sbjct 261 SIVDQTWGPSGTTQ-GLYADLSTATAA-----TINQLRQSFQIQKLLERDARGGTRYTEI 314
Query 503 MQGRWDIDIRFDELLMPEFIGGISRELSMRTVEQTVDQQSTSSQGQYAEALGSKSGIA-G 561
++ + + L PE++GG S +S+ + Q TS GQ +A + A G
Sbjct 315 LRSHFGVTSPDARLQRPEYLGGGSTLVSISPIAQ------TSGTGQTGQATPQGNLAAMG 368
Query 562 VYGSTSNNIEVFCDEESYIIGLLTVTPVPIYTQMLSKDFLYNGLLDHYQPEFDRIGFQPI 621
Y + ++ E YIIG+++V Y Q L + + + D+Y P F +G Q I
Sbjct 369 TYLAQNHGFTHSFVEHGYIIGVVSVRADLTYQQGLRRHWSRSTRYDYYFPAFAMLGEQAI 428
Query 622 TYKEVCPLNLGVADTVNKANQTFGYQRPWYEYVAKYDSAHGLFRTNMKNFV----MSRVF 677
KE+ N N FGYQ W EY GLFR+ + ++ F
Sbjct 429 LNKEI------YVTGSNSDNNVFGYQERWAEYRYNPSEITGLFRSTAAGTIDPWHYAQRF 482
Query 678 SGLPQLGQQFLLVDPDTVNQVFSVTEYTDKIFGY-VKFNATARLPISRVAIPRL 730
+ LP L F+ +P + + + F FN A P+ ++P L
Sbjct 483 TSLPTLNSTFIQDNPPLARNLAVGSAANGQQFLLDAFFNINAARPLPMYSVPGL 536
Score = 47.4 bits (111), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 43/152 (28%), Positives = 66/152 (43%), Gaps = 15/152 (10%)
Query 7 DATFDANN-----RIDVNSFDWSHVNNLTTDF--GRITPVFCELVPAKGSLRINPEFGLE 59
+ + DA+N R D+ +S L T F G + P+ CE + + +N
Sbjct 4 NKSVDAHNFAMVPRADIPRSRFSMQKTLKTTFDSGLLVPIMCEEILPGDTFNVNVTMFGR 63
Query 60 LMPMVFPVQTRMFARLNFFKVTLRSMWEDYSDFIS---NFRDDLEEPYILPGSQNFTTML 116
L +FPV + FF V R +W ++ F+ N D + Y +P +
Sbjct 64 LATPLFPVMDNLHLDSFFFFVPNRLVWTNWVKFMGEQDNPSDSIS--YTIPQQVSPAGGY 121
Query 117 GTGSLGDYLGVPTRN---AGAEVSLMAHPSQC 145
GSL DYLG+PT AG+ VS A P++
Sbjct 122 AIGSLQDYLGLPTVGQVTAGSTVSHSALPTRA 153
> Gokush_Human_gut_36_019_Microviridae_AG0279_putative.VP1
Length=582
Score = 102 bits (255), Expect = 1e-24, Method: Compositional matrix adjust.
Identities = 116/474 (24%), Positives = 186/474 (39%), Gaps = 72/474 (15%)
Query 303 YETTPFATQNKPANLRLLAYRFRAYESVYNAYYRDIRNNPFIVDGRPVYNKWLPSMKGGA 362
Y P PAN + A FRAY +YN ++RD ++D PV + G
Sbjct 131 YMGLPTGVSLDPANTPINALPFRAYNLIYNEWFRDEN----LIDSIPVL-----TTDGPD 181
Query 363 DNTTYSLHQCNWERDFLTTAVPNPQQG--------ANAPLVGLTVGDVVTRADDGTYSIQ 414
+ Y+L + D+ T+A+P PQ+G NAP+VG G D +YS
Sbjct 182 PVSNYTLRKRAKRHDYFTSALPWPQKGPSVDVGLTGNAPVVGFGDGQTWNFMSDTSYS-G 240
Query 415 KQTVLVDEDGSKYGISYKVSEDGESLVGVDYDPVSEKTPVTAI---------NSYAELAA 465
Q VL + + +V + + P+ ++T + +S E+ +
Sbjct 241 NQAVLGNPTDVLDNVGLQVFTNRKQFSTATMIPIMQETSRSGRWVNIGNQDQSSGTEIQS 300
Query 466 LTTTEGSGF-------------------------TIETLRYVNAYQKFLELNMRKGFSYK 500
G GF TI LR QKF E R G Y
Sbjct 301 TRAIRGDGFYFPNGILSNSLGQQPYADLSGVSAITINDLRQAFQIQKFYEKWARGGSRYT 360
Query 501 QIMQGRWDIDIRFDELLMPEFIGGISRELSMRTVEQTVDQQSTSSQGQYAEALGSKSGIA 560
+ ++ +++ L PE++GG +++ QT S S Q S
Sbjct 361 ETLRVMFNVISPDARLQRPEYLGGTHSRVNVVPTAQTSSTDSVSPQ--------SNLSAF 412
Query 561 GVYGSTSNNIEVFCDEESYIIGLLTVTPVPIYTQMLSKDFLYNGLLDHYQPEFDRIGFQP 620
GV G +++ E Y+IGL+ + Y Q L++ + L D Y P +G Q
Sbjct 413 GVLGDSAHGFNKSFVEHGYVIGLVCLRADITYQQGLNRMWSRRQLFDFYWPTLAHLGEQV 472
Query 621 ITYKEVCPLNLGVADTVNKANQTFGYQRPWYEYVAKYDSAHGLFRT----NMKNFVMSRV 676
+ KE+ G AD N FGYQ + EY K G R+ + + +++
Sbjct 473 VYNKEI--YTQGTADD----NGVFGYQERYAEYRYKPSMITGKLRSTDPQTLDVWHLAQK 526
Query 677 FSGLPQLGQQFLLVDPDTVNQVFSVTEYTDKIFGYVKFNATARLPISRVAIPRL 730
F LP+L Q F+ +P +N+V +V + + F F+ P+ ++P L
Sbjct 527 FDTLPKLNQDFIEENP-PINRVIAV-QNEPQFFADFWFDLKTSRPMPVYSVPGL 578
Score = 44.3 bits (103), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 29/117 (25%), Positives = 53/117 (45%), Gaps = 0/117 (0%)
Query 13 NNRIDVNSFDWSHVNNLTTDFGRITPVFCELVPAKGSLRINPEFGLELMPMVFPVQTRMF 72
N+ I + FD SH T D G + P F + V + ++ + + ++ P +F
Sbjct 20 NSPIQRSVFDRSHDYKTTLDSGYLIPFFVDEVLPGDTFKLRVNAFVRMNTLIAPFMDNVF 79
Query 73 ARLNFFKVTLRSMWEDYSDFISNFRDDLEEPYILPGSQNFTTMLGTGSLGDYLGVPT 129
FF V R +W+++ F ++ + L S + T G++ DY+G+PT
Sbjct 80 MDSFFFFVPTRLVWDNWQRFCGEKKNPGDSTDFLIPSLSGTNTFANGTIFDYMGLPT 136
> Gokush_Human_gut_37_015_Microviridae_AG033_putative.VP1
Length=582
Score = 102 bits (255), Expect = 1e-24, Method: Compositional matrix adjust.
Identities = 116/474 (24%), Positives = 186/474 (39%), Gaps = 72/474 (15%)
Query 303 YETTPFATQNKPANLRLLAYRFRAYESVYNAYYRDIRNNPFIVDGRPVYNKWLPSMKGGA 362
Y P PAN + A FRAY +YN ++RD ++D PV + G
Sbjct 131 YMGLPTGVSLDPANTPINALPFRAYNLIYNEWFRDEN----LIDSIPVL-----TTDGPD 181
Query 363 DNTTYSLHQCNWERDFLTTAVPNPQQG--------ANAPLVGLTVGDVVTRADDGTYSIQ 414
+ Y+L + D+ T+A+P PQ+G NAP+VG G D +YS
Sbjct 182 PVSNYTLRKRAKRHDYFTSALPWPQKGPSVDVGLTGNAPVVGFGDGQTWNFMSDTSYS-G 240
Query 415 KQTVLVDEDGSKYGISYKVSEDGESLVGVDYDPVSEKTPVTAI---------NSYAELAA 465
Q VL + + +V + + P+ ++T + +S E+ +
Sbjct 241 NQAVLGNPTDVLDNVGLQVFTNRKQFSTATMIPIMQETSRSGRWANIGNQDQSSGTEIQS 300
Query 466 LTTTEGSGF-------------------------TIETLRYVNAYQKFLELNMRKGFSYK 500
G GF TI LR QKF E R G Y
Sbjct 301 TRAIRGDGFYFPNGILSNSLGQQPYADLSGVSAITINDLRQAFQIQKFYEKWARGGSRYT 360
Query 501 QIMQGRWDIDIRFDELLMPEFIGGISRELSMRTVEQTVDQQSTSSQGQYAEALGSKSGIA 560
+ ++ +++ L PE++GG +++ QT S S Q S
Sbjct 361 ETLRVMFNVISPDARLQRPEYLGGTHSRVNVVPTAQTSSTDSVSPQ--------SNLSAF 412
Query 561 GVYGSTSNNIEVFCDEESYIIGLLTVTPVPIYTQMLSKDFLYNGLLDHYQPEFDRIGFQP 620
GV G +++ E Y+IGL+ + Y Q L++ + L D Y P +G Q
Sbjct 413 GVLGDSAHGFNKSFVEHGYVIGLVCLRADITYQQGLNRMWSRRQLFDFYWPTLAHLGEQV 472
Query 621 ITYKEVCPLNLGVADTVNKANQTFGYQRPWYEYVAKYDSAHGLFRT----NMKNFVMSRV 676
+ KE+ G AD N FGYQ + EY K G R+ + + +++
Sbjct 473 VYNKEI--YTQGTADD----NGVFGYQERYAEYRYKPSMITGKLRSTDPQTLDVWHLAQK 526
Query 677 FSGLPQLGQQFLLVDPDTVNQVFSVTEYTDKIFGYVKFNATARLPISRVAIPRL 730
F LP+L Q F+ +P +N+V +V + + F F+ P+ ++P L
Sbjct 527 FDTLPKLNQDFIEENP-PINRVIAV-QNEPQFFADFWFDLKTSRPMPVYSVPGL 578
Score = 44.3 bits (103), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 29/117 (25%), Positives = 53/117 (45%), Gaps = 0/117 (0%)
Query 13 NNRIDVNSFDWSHVNNLTTDFGRITPVFCELVPAKGSLRINPEFGLELMPMVFPVQTRMF 72
N+ I + FD SH T D G + P F + V + ++ + + ++ P +F
Sbjct 20 NSPIQRSVFDRSHDYKTTLDSGYLIPFFVDEVLPGDTFKLRVNAFVRMNTLIAPFMDNVF 79
Query 73 ARLNFFKVTLRSMWEDYSDFISNFRDDLEEPYILPGSQNFTTMLGTGSLGDYLGVPT 129
FF V R +W+++ F ++ + L S + T G++ DY+G+PT
Sbjct 80 MDSFFFFVPTRLVWDNWQRFCGEKKNPGDSTDFLIPSLSGTNTFANGTIFDYMGLPT 136
Lambda K H a alpha
0.318 0.133 0.393 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 68431296