bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: Microviridae_proteins.fasta
575 sequences; 147,441 total letters
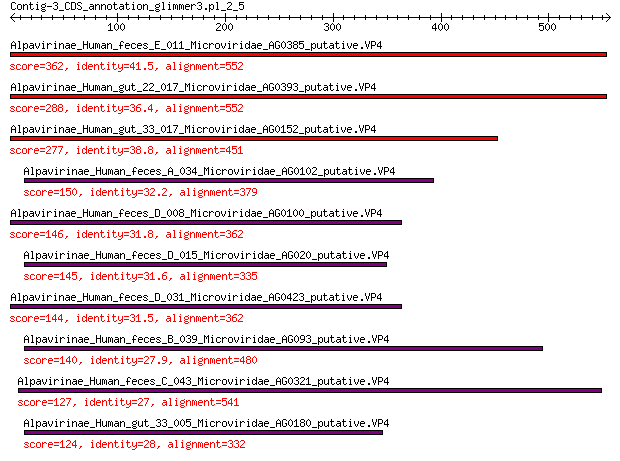
Query= Contig-3_CDS_annotation_glimmer3.pl_2_5
Length=556
Score E
Sequences producing significant alignments: (Bits) Value
Alpavirinae_Human_feces_E_011_Microviridae_AG0385_putative.VP4 362 1e-118
Alpavirinae_Human_gut_22_017_Microviridae_AG0393_putative.VP4 288 7e-91
Alpavirinae_Human_gut_33_017_Microviridae_AG0152_putative.VP4 277 1e-86
Alpavirinae_Human_feces_A_034_Microviridae_AG0102_putative.VP4 150 7e-41
Alpavirinae_Human_feces_D_008_Microviridae_AG0100_putative.VP4 146 9e-40
Alpavirinae_Human_feces_D_015_Microviridae_AG020_putative.VP4 145 3e-39
Alpavirinae_Human_feces_D_031_Microviridae_AG0423_putative.VP4 144 4e-39
Alpavirinae_Human_feces_B_039_Microviridae_AG093_putative.VP4 140 8e-38
Alpavirinae_Human_feces_C_043_Microviridae_AG0321_putative.VP4 127 4e-33
Alpavirinae_Human_gut_33_005_Microviridae_AG0180_putative.VP4 124 5e-32
> Alpavirinae_Human_feces_E_011_Microviridae_AG0385_putative.VP4
Length=611
Score = 362 bits (928), Expect = 1e-118, Method: Compositional matrix adjust.
Identities = 229/610 (38%), Positives = 339/610 (56%), Gaps = 60/610 (10%)
Query 1 MTEKER--EKYLATECFHPRKVTNRYTHETLFVRCGTCPSCLVHRSNIQCALISNMSSHF 58
M KE KYL TEC P+++ N Y+H+ +F CG C SC++++SN A NM++HF
Sbjct 1 MIPKEELLTKYLFTECLRPQRIVNPYSHDVIFAPCGHCKSCIMNKSNFATAYAMNMATHF 60
Query 59 KHAYFFTLTYSDEFVPRVSLEVVERCDA----ESEIDAYMSDSDPRHLP----------Y 104
K+ YF TLTY D F+P +S+EVV R + + +S SDPR L
Sbjct 61 KYCYFVTLTYKDIFLPYLSVEVVRRSGNRYLFDENFETMVSTSDPRFLTPEYYHDRDFSL 120
Query 105 DDSRYQIAATY------LPRSGCFRVHDSGRVRDFSETEDSYQFLHTFSGKEIRDLLVSS 158
D ++ ++ + +PR + S R R F + + +F E++D+L+ +
Sbjct 121 DSAQNEVEQVFDVGFQSIPRDVSVKSKGSFRFRSFDD--EPLKFCIPMKLTELQDILIKA 178
Query 159 NGRYDFSRKCVVFPSIDECRNEILVLNPYDQNLFFKRLRKLISER--YDEKICYYLVSEY 216
NGRYD+ +K VV+PS+ +C+ +I VL D LFFKRLR+ + KICYY+VSEY
Sbjct 179 NGRYDYGKKKVVYPSLADCKLQIPVLQSRDIELFFKRLRRNLDSHGFTSSKICYYVVSEY 238
Query 217 GGRTYRPHWHGILFFNSDALTSSICELVSKSWSYGRTDCSLSRGSAAGYVASYINSFVDL 276
G +TYRPHWH +LFFNS+ +T ++ E +SK+WSYGR D SLSRG+AA YVA+Y+NS L
Sbjct 239 GPQTYRPHWHCLLFFNSEEITQTLREDISKAWSYGRIDYSLSRGAAASYVAAYVNSAACL 298
Query 277 PDFFNRHKEIKPKSYHSKGLSVNSLFSRTSDISQVDEVAASCFDGFSVPINGEYVTVKPS 336
P + KEI+P+S+HSKG N +F ++SD+S++ +++ FDG +V NG+ V ++P
Sbjct 299 PFLYVGQKEIRPRSFHSKGFGSNKVFPKSSDVSEISKISDLFFDGVNVDSNGKVVNIRPV 358
Query 337 RSYEHTVFPRISDPFFKDPHSCVDLFFGAFTASNRLIRDGYISID------ENLSVWQLS 390
RS E VFPR S+ FF D +C LF RL+ GY+ ID ++ + L
Sbjct 359 RSCELAVFPRFSNDFFSDSDTCCKLFQSVIETPERLVSRGYLGIDTPNFGSDDFRLSDLV 418
Query 391 RRYAEYY--------FSVR--AGHYNVSYYDSLIFKFTRLETDQI-DFDCITGKIYRFFS 439
R Y+EYY FS+R G+ Y D LIF+ RL + + D I G++YR F+
Sbjct 419 RAYSEYYERNFTDFSFSLRFLRGYRTRDYADELIFREARLFDGYVFNKDMIFGRLYRLFA 478
Query 440 AVSRTLRFWHLDCYVLPEDlkksllklfissVDYWSKKELKFINDFYDYLSA-HPESDSF 498
V R RFW+L Y LK ++ K++ +YW KKE +F+ +++YL + + F
Sbjct 479 KVLRCFRFWNLKQYTDSWSLKVAIKKIWSYGWEYWKKKEYRFLTTYFEYLEGCNDDERLF 538
Query 499 LKSRTVG----------FVCPPKENI------SFWNDLNTTLSSLKNSVTQRIFQKVKHK 542
L RT G + +E+ F+ TL L + + KVKHK
Sbjct 539 LLVRTSGSGLATDSPHSWTYTQREDYVNCLSDDFYKRYMNTLKWLTARTEKVLKDKVKHK 598
Query 543 NYNDISGLLF 552
+ND+ G+L
Sbjct 599 EFNDMQGVLL 608
> Alpavirinae_Human_gut_22_017_Microviridae_AG0393_putative.VP4
Length=562
Score = 288 bits (737), Expect = 7e-91, Method: Compositional matrix adjust.
Identities = 201/591 (34%), Positives = 297/591 (50%), Gaps = 68/591 (12%)
Query 1 MTEKEREKYLATECFHPRKVTNRYTHETLFVRCGTCPSCLVHRSNIQCALISNMSSHFKH 60
M KE + L T C HPR V N+YTHE + V CG CPSC++ RS++Q L++ S+ F++
Sbjct 1 MITKELQNKLVTRCQHPRTVVNKYTHEPVVVSCGHCPSCILRRSSVQTNLLTTYSAQFRY 60
Query 61 AYFFTLTYSDEFVPRVSLEVVERCDAESEIDAYMSDSDPRHLPYDDSRYQIAATYLPRSG 120
YF TLTY+ F+P + + VVE C + + + + D D + Y +PRS
Sbjct 61 VYFVTLTYAPSFLPTLEVSVVETCTDDIADVSCVPNIDELDAS-DPNTYLFGFRSVPRSS 119
Query 121 CFRVHDSGRVRDFSETEDSYQFLHTFSGKEIRDLLVSSNGRYDFSRKCVVFPSIDECRNE 180
++ S R F + + +F + KE+ +L N N
Sbjct 120 SVKLKSSTVERTFKDPD--VKFSYPMKPKELLSILGKIN---------------HNVPNR 162
Query 181 ILVLNPYDQNLFFKRLRKLISERYDEKICYYLVSEYGGRTYRPHWHGILFFNSDALTSSI 240
I + D +LF KRLR S DEK+ YY VSEYG ++RPHWH +LF NS+ + ++
Sbjct 163 IPYICNRDLDLFLKRLR---SYYLDEKLRYYAVSEYGPTSFRPHWHLLLFSNSERFSRTV 219
Query 241 CELVSKSWSYGRTDCSLSRGSAAGYVASYINSFVDLPDFFNRH-KEIKPKSYHSKGLSVN 299
CE VSK+WSYGR D SLSRG AA YVASY+NSFV LPDF+ + K ++PKS+HS G + +
Sbjct 220 CENVSKAWSYGRCDASLSRGFAAPYVASYVNSFVALPDFYTQMPKVVRPKSFHSIGFTES 279
Query 300 SLFSRTSDISQVDEVAASCFDGFSVPINGEYVTVKPSRSYEHTVFPRISDPFFKDPHSCV 359
+LF R +++VDE+ C +G V +G + T+KP+ Y +FPR SD K P S
Sbjct 280 NLFPRKVRVAEVDEITDKCLNGVRVERDGYFRTIKPTWPYLLRLFPRFSDAIRKSPSSIY 339
Query 360 DLFFGAFTASNRLIRDGYISI-------DENLSVWQLSRRYAEYY--FSVRAGHYN---- 406
L F AFTA R+IR G I S+ ++Y Y + H N
Sbjct 340 QLLFAAFTAPERVIRSGCADIGCDPFGESSKQSILSFCKQYLNYVDNYGKSNEHRNFLSP 399
Query 407 ---VSYYDSLIFKFTRLETDQIDFDCI--TGKIYRFFSAVSRTLRFWHLD-CYVL----- 455
+ + D LI RL D +D + ++YRFF +S+ +R + D C L
Sbjct 400 QASLPHSDVLILSECRL-YDGVDLETTHRVSRVYRFFLGISKFIRTYSTDGCSELFWSSG 458
Query 456 -PED----lkksllklfissVDYWSKKELKFINDFYDYLSAHPESD---------SFLKS 501
P ++ L + V++W++ E + DFY L + + SF +
Sbjct 459 TPGGDLFCRERFLRIISEKIVNFWNRYEYNRLVDFYQTLEDSNDKELVDFELRNYSFRYN 518
Query 502 RTVGFVCPPKENISFWNDLNTTLSSLKNSVTQRIFQKVKHKNYNDISGLLF 552
R+V ++N +N+L + L + + KVKHK ND+SG+
Sbjct 519 RSV------RDNERPYNEL-PLVRRLAAASLMKCRDKVKHKKVNDLSGIFL 562
> Alpavirinae_Human_gut_33_017_Microviridae_AG0152_putative.VP4
Length=565
Score = 277 bits (708), Expect = 1e-86, Method: Compositional matrix adjust.
Identities = 175/471 (37%), Positives = 247/471 (52%), Gaps = 43/471 (9%)
Query 1 MTEKEREKYLATECFHPRKVTNRYTHETLFVRCGTCPSCLVHRSNIQCALISNMSSHFKH 60
M KE + L T C +PR V N+YTHE + V CG CPSC++ RS IQ L++ S+ F++
Sbjct 1 MITKELQNKLVTRCQNPRTVVNKYTHEPVVVSCGACPSCVLRRSGIQTNLLTTYSAQFRY 60
Query 61 AYFFTLTYSDEFVPRVSLEVVERCDAESEIDAYMSDSDPRHL-PYDDSRYQIAATYLPRS 119
YF TLTY+ F+P + + V+E C +I S D L P D++RY +PRS
Sbjct 61 VYFVTLTYAPCFLPTLEVSVIETC--TDDIADVSSVPDINDLDPCDNNRYLFGFCSVPRS 118
Query 120 GCFRVHDSGRVRDFSETEDSYQFLHTFSGKEIRDLLVSSNGRYDFSRKCVVFPSIDECRN 179
++ +S R F + E +F + KE+ +L N N
Sbjct 119 ASVKLKNSTVERTFKDPE--VRFSYPMKPKELLSILDKIN---------------HNVPN 161
Query 180 EILVLNPYDQNLFFKRLRKLISERYDEKICYYLVSEYGGRTYRPHWHGILFFNSDALTSS 239
I + D +LF KRLR EK+ YY VSEYG +YRPHWH +LF NS+ + +
Sbjct 162 RIPYICNRDLDLFLKRLRSYYPY---EKLRYYAVSEYGPTSYRPHWHLLLFSNSEQFSKT 218
Query 240 ICELVSKSWSYGRTDCSLSRGSAAGYVASYINSFVDLPDFFNRHKE-IKPKSYHSKGLSV 298
I E VSK+WSYGR D SLSRG AA YVASY+NSFV LP F+ ++PKS+HS G +
Sbjct 219 ILENVSKAWSYGRCDASLSRGFAAPYVASYVNSFVALPSFYTEMPRFLRPKSFHSIGFTE 278
Query 299 NSLFSRTSDISQVDEVAASCFDGFSVPINGEYVTVKPSRSYEHTVFPRISDPFFKDPHSC 358
++LF R IS++DEV C +G V +G + +KPS Y +FPR SD K P S
Sbjct 279 SNLFPRKVRISEIDEVTDKCLNGVRVERDGYFRILKPSWPYLLRLFPRFSDAIRKSPSSI 338
Query 359 VDLFFGAFTASNRLIRDGYISI-------DENLSVWQLSRRYAEY---------YFSVRA 402
L F AFTA R+IR G I + S+ ++Y Y + + +
Sbjct 339 YQLLFAAFTAPARVIRSGCADIGCDPFVENSKQSLLSFCKQYLNYVDNHGKSNEFKNFLS 398
Query 403 GHYNVSYYDSLIFKFTRLETDQIDFDCI--TGKIYRFFSAVSRTLRFWHLD 451
H ++ + D LI RL D +D + ++YRFF +++ +R + D
Sbjct 399 PHADLPHSDVLILTECRL-YDGVDLEAAHRLSRVYRFFLGIAKFIRTYSTD 448
> Alpavirinae_Human_feces_A_034_Microviridae_AG0102_putative.VP4
Length=539
Score = 150 bits (378), Expect = 7e-41, Method: Compositional matrix adjust.
Identities = 122/389 (31%), Positives = 184/389 (47%), Gaps = 48/389 (12%)
Query 14 CFHPRKVTNRYTHETLFVRCGTCPSCLVHRSNIQCALISNMS-SHFKHAYFFTLTYSDEF 72
C HPR + N+YT + ++V CGTC C VH I+ L N+ + ++ F TLTYS E+
Sbjct 12 CLHPRVIKNKYTGDPIYVPCGTCEFC-VHNKAIKAELKCNVQLASSRYCEFITLTYSTEY 70
Query 73 VPRVSLEVVERCDAESEIDAYMSDSDPRHLPYDDSRYQIAATYLPRSGCFRVHD-SGRVR 131
+P Y R ++ LPR + G R
Sbjct 71 LPVGKF-------------------------YQGPRGEVRFCSLPRDFVYSYKTVQGYTR 105
Query 132 DFSETEDSYQFLHTFSGKEIRDLLVSSNGRYDFSRKCVVFPSIDECRN----EILV--LN 185
+ S + ++ F S + L ++ Y FP N E LV LN
Sbjct 106 NISFNDKTFDFDTQLSWSSAQLLQKKAHLHY------TSFPDGRCIYNRPYMEGLVGYLN 159
Query 186 PYDQNLFFKRLRKLISERYDEKICYYLVSEYGGRTYRPHWHGILFFNSDALTSSICELVS 245
+D LFFKRL + I +EKI YY+V EYG T+RPH+H +LF +S L SI + VS
Sbjct 160 YHDIQLFFKRLNQNIRRITNEKIYYYVVGEYGPTTFRPHFHILLFHDSRKLRESIRQFVS 219
Query 246 KSWSYGRTDCSLSRGSAAGYVASYINSFVDLPDFFNRHKEIKPKSYHSKGLSVNSLFSRT 305
KSW +G +D SA+ YVA Y+NS LPDFF + IKP S + S F+
Sbjct 220 KSWRFGDSDTQPVWSSASCYVAGYVNSTACLPDFFKNSRHIKPFGRFSVNFA-ESAFNEV 278
Query 306 SDISQVDEVAASCFDGFSVPINGEYVTVKPSRSYEHTVFPRISDPFFKDPHSCV--DLFF 363
+ +E+ + +DG + +NG+ V+P RS+ + ++PR+ K H+ V D+
Sbjct 279 FKPEENEEIFSLFYDGRVLNLNGKPTIVRPKRSHINRLYPRLD----KSKHATVVDDIRI 334
Query 364 GAFTASNRLIRDGYISIDENLSVWQLSRR 392
+ + + + IDE +S ++LS+R
Sbjct 335 ASVVSRLPQVVAKFGFIDE-VSDFELSKR 362
> Alpavirinae_Human_feces_D_008_Microviridae_AG0100_putative.VP4
Length=532
Score = 146 bits (369), Expect = 9e-40, Method: Compositional matrix adjust.
Identities = 115/368 (31%), Positives = 178/368 (48%), Gaps = 37/368 (10%)
Query 1 MTEKEREKYLATECFHPRKVTNRYTHETLFVRCGTCPSCLVHRSNIQCALISNMSSHFKH 60
M ++E + + + C H + N YT E + V CGTCP+C ++S + + S +H
Sbjct 1 MNQQEFIQKVFSMCQHQVQTKNPYTGELIMVPCGTCPACRFNKSILSQNKVHAQSLISRH 60
Query 61 AYFFTLTYSDEFVPRVSLEVVERCDAE-SEIDAYMSDSDPRHLPYDDSRYQIAATYLPRS 119
YF TLTY+ ++P E+ E DA+ I A+ D +P + Y TY
Sbjct 61 VYFVTLTYAQRYIPYYEYEI-EALDADFLAITAHCRDRNPMYRTY---------TYRGTK 110
Query 120 GCFRVHD--SGRVRDFSETEDSYQFLHTFSGKEIRDLLVSSNGRYDFSRKCVVFPSIDEC 177
R+ S +V+ FS + + + +++ K +S +G+Y P++ +
Sbjct 111 HKLRIRGLASPKVKSFSFSVNR-DYWTSYAQKAN----LSFDGKY---------PALSD- 155
Query 178 RNEILVLNPYDQNLFFKRLRKLISE-RYDEKICYYLVSEYGGRTYRPHWHGILFFNSDAL 236
I L D +L+ KR+RK IS+ +E I Y+V EYG T+RPH+H +LFFNSD L
Sbjct 156 --RIPYLLHDDVSLYMKRVRKYISKLGINETIHTYVVGEYGPVTFRPHFHLLLFFNSDEL 213
Query 237 TSSICELVSKSWSYGRTDCSLSRGSAAGYVASYINSFVDLPDFFNRHKEIKPKSYHSK-- 294
SI + W +GR DCS SRG A YV+SY+NSF +P + I+P + S
Sbjct 214 AQSIVRIARSCWRFGRVDCSASRGDAEDYVSSYLNSFSSIPLHIQEIRAIRPFARFSNKF 273
Query 295 GLSVNSLFSRTSDISQVDEVAASCFDGFSVPINGEYVTVKPSRSYEHTVFPRISDPFFKD 354
G S + + DE+ +G S+P NG T+ P R+ T F R + D
Sbjct 274 GFSFFESSIKKAQSGDFDEI----LNGKSLPYNGFNTTIFPWRAIIDTCFYRPALRRRSD 329
Query 355 PHSCVDLF 362
H ++
Sbjct 330 IHELTEIL 337
> Alpavirinae_Human_feces_D_015_Microviridae_AG020_putative.VP4
Length=563
Score = 145 bits (365), Expect = 3e-39, Method: Compositional matrix adjust.
Identities = 106/372 (28%), Positives = 173/372 (47%), Gaps = 47/372 (13%)
Query 14 CFHPRKVTNRYTHETLFVRCGTCPSCLVHRSNIQCALISNMSSHFKHAYFFTLTYSDEFV 73
C HP+ + N+YT + + V CG CP CL+ +++ + + ++ YF TLTY+ ++V
Sbjct 12 CEHPQVIQNKYTGDYVKVDCGQCPYCLIKKADRSTQKCDFVKYNHRYCYFVTLTYNTQYV 71
Query 74 PRVSLEVVE---------------------RCDAESEIDAYMSD--SDPRHLPY------ 104
P++SL +E R +S ++ + D S + PY
Sbjct 72 PKMSLTQIEDYLSEWLPVRPPKSFGTQLTARMLTDSRVNKKIPDFMSAEVNRPYMLEHLR 131
Query 105 --DDSRYQIAATYLP----RSGCFRVHDSGRVRDFSETEDSYQFLHTFSGKEIRDLLVSS 158
+ RY+ A P ++ + + RV +D Y + EI + L
Sbjct 132 LLEADRYKALALRYPNFISKARPYILRSIPRVSKLQNFKDEYFEELVWMLPEIAESLKKK 191
Query 159 NGRYDFSRKCVVFPSIDECRNEILVLNPYDQNLFFKRLRKLISERYD--EKICYYLVSEY 216
N + FP + + + +N D LF KRLRK +S++ EKI Y+VSEY
Sbjct 192 NN----TDANGAFP---QFKGLLKYVNIRDYQLFAKRLRKYLSKKVGKYEKIHSYVVSEY 244
Query 217 GGRTYRPHWHGILFFNSDALTSSICELVSKSWSYGRTDCSLSRGSAAGYVASYINSFVDL 276
+T+RPH+H + FF+SD + + + V +SW GR D L+R A YV++Y+NS V +
Sbjct 245 SPKTFRPHFHILFFFDSDEIAKNFRQAVYQSWRLGRVDTQLAREQANSYVSNYLNSVVSI 304
Query 277 PDFFNRHKEIKPKSYHSKGLSVNSLFSRTSDISQVDEVAASCFDGFSVPINGEYVTVKPS 336
P + K I+P+S S + I + + A+ FDG S N ++V PS
Sbjct 305 PFVYKAKKSIRPRSRFSNLFGFEEV---KEGIRKAQDKRAALFDGLSYISNQKFVRYVPS 361
Query 337 RSYEHTVFPRIS 348
S +FPR +
Sbjct 362 GSLIDRLFPRFT 373
> Alpavirinae_Human_feces_D_031_Microviridae_AG0423_putative.VP4
Length=532
Score = 144 bits (363), Expect = 4e-39, Method: Compositional matrix adjust.
Identities = 114/368 (31%), Positives = 177/368 (48%), Gaps = 37/368 (10%)
Query 1 MTEKEREKYLATECFHPRKVTNRYTHETLFVRCGTCPSCLVHRSNIQCALISNMSSHFKH 60
M ++E + + + C H + N YT E + V CGTCP+C ++S + + S +H
Sbjct 1 MNQQEFIQKVFSMCQHQVQTKNPYTGELIMVPCGTCPACRFNKSILSQNKVHAQSLVSRH 60
Query 61 AYFFTLTYSDEFVPRVSLEVVERCDAE-SEIDAYMSDSDPRHLPYDDSRYQIAATYLPRS 119
YF TLTY+ ++P E+ E DA+ I A+ D +P + Y TY
Sbjct 61 VYFVTLTYAQRYIPYYEYEI-EALDADFLAITAHCCDRNPMYRTY---------TYRGTK 110
Query 120 GCFRVHD--SGRVRDFSETEDSYQFLHTFSGKEIRDLLVSSNGRYDFSRKCVVFPSIDEC 177
R+ S V+ FS + + + +++ K +S NG+Y P++
Sbjct 111 HKLRIRGLASPNVKSFSFSVNR-DYWTSYAQKAN----LSFNGKY---------PALS-- 154
Query 178 RNEILVLNPYDQNLFFKRLRKLISE-RYDEKICYYLVSEYGGRTYRPHWHGILFFNSDAL 236
I L D +L+ KR+RK IS+ +E I Y+V EYG ++RPH+H +LFF+SD L
Sbjct 155 -GRIPYLLHGDVSLYMKRVRKYISKLGINETIHTYIVGEYGPSSFRPHFHLLLFFDSDEL 213
Query 237 TSSICELVSKSWSYGRTDCSLSRGSAAGYVASYINSFVDLPDFFNRHKEIKPKSYHSK-- 294
+I + S W +GR DCS SRG A YV+SY+NSF +P + I+P + S
Sbjct 214 AQNIIRIASSCWRFGRVDCSASRGDAEDYVSSYLNSFSSIPLHIQEIRAIRPFARFSNKF 273
Query 295 GLSVNSLFSRTSDISQVDEVAASCFDGFSVPINGEYVTVKPSRSYEHTVFPRISDPFFKD 354
G S + + DE+ +G S+P NG T+ P R+ T F R + D
Sbjct 274 GYSFFESSIKKAQSGNFDEI----LNGKSLPYNGFDTTIFPWRAIIDTCFYRPALRRHSD 329
Query 355 PHSCVDLF 362
H ++
Sbjct 330 IHELTEIL 337
> Alpavirinae_Human_feces_B_039_Microviridae_AG093_putative.VP4
Length=559
Score = 140 bits (354), Expect = 8e-38, Method: Compositional matrix adjust.
Identities = 134/512 (26%), Positives = 228/512 (45%), Gaps = 59/512 (12%)
Query 14 CFHPRKVTNRYTHETLFVRCGTCPSCLVHRSNIQCALISNMSSHFKHAYFFTLTYSDEFV 73
C H +TNRY + V CG C C+ R+ + S FK++YF TLTY +E +
Sbjct 14 CQHRSFITNRYNGARITVDCGQCDYCIHKRAQKASMRVKTAGSAFKYSYFVTLTYDNEHI 73
Query 74 PRVSLEVVERCDAESEIDAYMSDSDPRHLPYDDSRYQIAATYLPRSGCFRVHDSGRVRD- 132
P ++ +V+ SE + + S H + D ++ Y+P S +R D+ +R
Sbjct 74 PLMNCKVLH-----SEYEDVVGISGDIH--FGDEYHK----YIPVSE-YRCDDNSMLRHI 121
Query 133 -FSETEDSYQFLHTFSGKEIRD---------LLVSSNGRYDFSRKCVV---FPSIDECRN 179
F + + + F +EI++ L +++ + + + V +P+ ++
Sbjct 122 FFEQVQGTVPF-----DREIKEYVPVKDNWFLSMAAIRSFIYKTQSVDKTDYPASEQYGR 176
Query 180 EILV--LNPYDQNLFFKRLRKLISERYD--EKICYYLVSEYGGRTYRPHWHGILFFNSDA 235
+ L+ LN D + KRLRK + ++ E +Y V EYG +RPH+H +LF NSD
Sbjct 177 DNLIPFLNYVDVQNYIKRLRKYLFQQLGKYETFHFYAVGEYGPVHFRPHYHLLLFTNSDK 236
Query 236 LTSSICELVSKSWSYGRTDCSLSRGSAAGYVASYINSFVDLPDFFNRHKEIKPKSYHSKG 295
++ + KSW GR+D S G A YVASY+NS P + + +PKS S G
Sbjct 237 VSEVLRYCHDKSWKLGRSDFQRSAGGAGSYVASYVNSLCSAPLLYRSCRAFRPKSRASVG 296
Query 296 LSVNSLFSRTSDI-------SQVDEVAASCFDGFSVPINGEYVTVKPSRSYEHTVFPRIS 348
F + D +Q+++ S +G NG V P SY T+ PR S
Sbjct 297 -----FFEKGCDFVEDDDPYAQIEQKIDSVVNGRVYNFNGVSVRSTPPLSYIRTLLPRFS 351
Query 349 DPFFKDPHSCVDLFFGAFTASNRLIRDGYISIDENLSVWQLSRRYAEYYFSVRAGHYNVS 408
D + + + + R+ R G+ ++ S+ L R Y +Y + ++
Sbjct 352 SARNDDGIAIARILYAVHSTPKRIARFGFADYKQD-SILSLVRTYYQYL----KVNPILT 406
Query 409 YYDSLIFKFTRLET------DQIDFDCITGKIYRFFSAVSRTLRFWHLDCYVLP-EDlkk 461
D LI +R T +D + K+YR F V + R WHL +
Sbjct 407 DDDKLILHASRCLTRFVNCSSDVDIESYINKLYRLFLYVYKFFRNWHLPFFGSDISAFSG 466
Query 462 sllklfissVDYWSKKELKFINDFYDYLSAHP 493
++ + + ++Y KK+ + + D ++ SA+P
Sbjct 467 RIMFIIKTGIEYEKKKDYESLRDVFNIRSANP 498
> Alpavirinae_Human_feces_C_043_Microviridae_AG0321_putative.VP4
Length=546
Score = 127 bits (318), Expect = 4e-33, Method: Compositional matrix adjust.
Identities = 146/563 (26%), Positives = 229/563 (41%), Gaps = 71/563 (13%)
Query 8 KYLATECFHPRKVTNRYTHETLFVRCGTCPSCLVHRSNIQCALISNMSSHFKHAYFFTLT 67
K L+ C H + V N+YT + ++ CG C +CL ++ + + +S K+ F TLT
Sbjct 9 KELSKTCLHGKNVYNKYTGQMMYQSCGKCEACLSRLASARSIKVGVQASLSKYVMFVTLT 68
Query 68 YSDEFVPRVSLEVVERCDAESEIDA-YMSDSDPR---HLPYDDSRYQIAATYLPRSGCFR 123
Y VP+ C S D Y+ PR + Y+ S Q L FR
Sbjct 69 YDTYHVPK--------CKIYSNGDNNYVLVVKPRVKDYFYYETSDGQKRLKGLSYDDDFR 120
Query 124 VHDSGRVRDFSETEDSYQFLHTFSGKEIRDLLVSSNGRYDFSRKCVVFPSIDECRNEILV 183
V +F +D ++ F + D+ G Y + + S +C+
Sbjct 121 V-------EFKADKD---YIENFKKQANLDV----KGCYPHLKDMYGYLSRKDCQ----- 161
Query 184 LNPYDQNLFFKRLRKLISERYDEKICYYLVSEYGGRTYRPHWHGILFFNSDALTSSICEL 243
LF KR+RK I DEKI Y+V EY + +RPH+H + FFNS+ L+ S +
Sbjct 162 -------LFMKRVRKQIRNYTDEKIHTYIVGEYSPKHFRPHFHILFFFNSNELSQSFGSI 214
Query 244 VSKSWSYGRTDCSLSRGSAAGYVASYINSFVDLPDFFNRHKEIKPKSYHSKGLSVNSLFS 303
V W +G D S SRG A YVA Y+NSF LP +I P S S S F
Sbjct 215 VRSCWKFGLVDWSQSRGDAESYVAGYVNSFARLPYHLGSSPKIAPFGRFSNHFS-ESCFD 273
Query 304 ------RTSDISQVDEVAASCFDGFSVPINGEYVTVKPSRSYEHTVFPRISDPFFKDPHS 357
R S Q V A +G + +NG+ + +PSRS + F R + H
Sbjct 274 DAKQTLRDSFFGQKTPVFAPFLNGVTNLVNGKLLLTRPSRSCVDSCFFRKARDGRLSSHE 333
Query 358 CVDLFFGAFTASNR----LIRDG--YISIDENLSVWQLSR--RYA--EYYFSVRAGHYNV 407
+ L + RDG Y ++ + + + RY E S+ + +
Sbjct 334 LLHLIRAVCHVVEQGKRFFARDGLNYTFLNHARFIVKACKSFRYQKREKLLSLPSRLVTL 393
Query 408 SYYDSLIFKFTRLETDQIDFD-CITGKIYRFFSAVSRTLRFWHLDCYVLPEDlkksllkl 466
YY + + D D + IYR R +RFW +D E ++ +
Sbjct 394 LYYSRVDLTKDIQDYKNPDVDEAMAQSIYRVLLYTMRFVRFWKIDKMPYHEGIRLLDMCR 453
Query 467 fissVDYWSKKELKFINDFYDYLSAHPESDSFLKSRTVGFVCPPKENISFWNDLNTTLSS 526
+Y+ + +++ + +L E + F P EN F + L T+ +
Sbjct 454 -----EYYRLLDYQYLRQRFQFLEQCEED-------LLDFYLHPTEN--FADYLPRTIVT 499
Query 527 LKNSVTQRIFQK-VKHKNYNDIS 548
N + +QK +KH+ ND++
Sbjct 500 GINQYSLERYQKAIKHREINDLN 522
> Alpavirinae_Human_gut_33_005_Microviridae_AG0180_putative.VP4
Length=536
Score = 124 bits (310), Expect = 5e-32, Method: Compositional matrix adjust.
Identities = 93/337 (28%), Positives = 158/337 (47%), Gaps = 36/337 (11%)
Query 14 CFHPRKVTNRYTHETLFVRCGTCPSCLVHRSNIQCALISNMS-SHFKHAYFFTLTYSDEF 72
C HP + N+YT + ++V CGTC CL +++ IQ L N+ + + +F TLTY+ E
Sbjct 12 CLHPVIIKNKYTGDPIYVECGTCEVCLSNKA-IQKELRCNIQLASSRCCFFVTLTYATEH 70
Query 73 VPRVSLEVVERCDAESEIDAYMSDSDPRHLPYDDSRYQIAATYLPRSGCFR-VHDSGRVR 131
+P + + +D HL +PR + V G R
Sbjct 71 IP---------------VARFYKLNDSYHL-----------CCVPRDHVYTYVTSQGYNR 104
Query 132 DFSETEDSYQFLHTFSGKEIRDLLVSSN---GRYDFSRKCVVFPSIDECRNEILVLNPYD 188
S ++ + + + + LL ++ Y R V +P++ + + LN D
Sbjct 105 KMSFCDEEFDYPTNLRDEAVTALLDKTHLDRTVYPDGRSVVKYPNMGDL---LPYLNYRD 161
Query 189 QNLFFKRLRKLISERYDEKICYYLVSEYGGRTYRPHWHGILFFNSDALTSSICELVSKSW 248
LF KR+ + I + DEKI Y V EYG +T+RPH+H + FF+S+ L +LV K+W
Sbjct 162 VQLFHKRINQQIKKYTDEKIYSYTVGEYGPKTFRPHFHLLFFFDSERLAQVFGQLVDKAW 221
Query 249 SYGRTDCSLSRGSAAGYVASYINSFVDLPDFFNRHKEIKPKSYHSKGLSVNSLFSRTSDI 308
+G +D SA+ YVA Y+NS LP+F+ +++I P S+ + F
Sbjct 222 RFGNSDTQRVWSSASSYVAGYLNSSHCLPEFYRCNRKIAPFGRFSQYFAERP-FIEAFKP 280
Query 309 SQVDEVAASCFDGFSVPINGEYVTVKPSRSYEHTVFP 345
+ ++V +G + + G+ +P RS + ++P
Sbjct 281 EENEKVFDKFVNGIYLSLGGKPTLCRPKRSLINRLYP 317
Lambda K H a alpha
0.323 0.137 0.428 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 50940078