bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: Microviridae_proteins.fasta
575 sequences; 147,441 total letters
Query= Contig-40_CDS_annotation_glimmer3.pl_2_2
Length=278
Score E
Sequences producing significant alignments: (Bits) Value
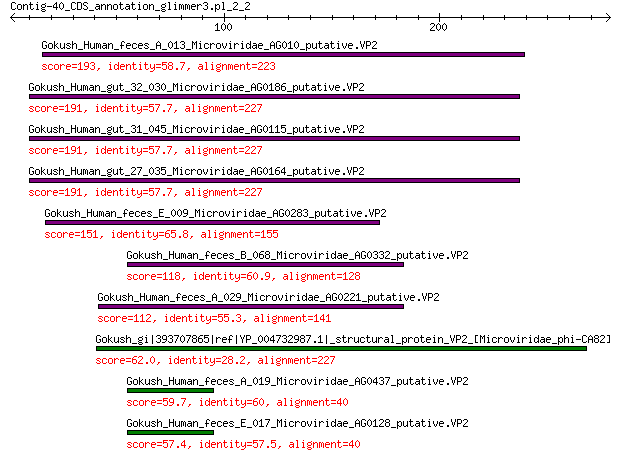
Gokush_Human_feces_A_013_Microviridae_AG010_putative.VP2 193 2e-61
Gokush_Human_gut_32_030_Microviridae_AG0186_putative.VP2 191 3e-60
Gokush_Human_gut_31_045_Microviridae_AG0115_putative.VP2 191 3e-60
Gokush_Human_gut_27_035_Microviridae_AG0164_putative.VP2 191 3e-60
Gokush_Human_feces_E_009_Microviridae_AG0283_putative.VP2 151 4e-45
Gokush_Human_feces_B_068_Microviridae_AG0332_putative.VP2 118 2e-32
Gokush_Human_feces_A_029_Microviridae_AG0221_putative.VP2 112 2e-30
Gokush_gi|393707865|ref|YP_004732987.1|_structural_protein_VP2_... 62.0 3e-13
Gokush_Human_feces_A_019_Microviridae_AG0437_putative.VP2 59.7 4e-12
Gokush_Human_feces_E_017_Microviridae_AG0128_putative.VP2 57.4 2e-11
> Gokush_Human_feces_A_013_Microviridae_AG010_putative.VP2
Length=276
Score = 193 bits (491), Expect = 2e-61, Method: Compositional matrix adjust.
Identities = 131/234 (56%), Positives = 165/234 (71%), Gaps = 22/234 (9%)
Query 16 VGNLDSALSRITRTASENTAKSAQMASEQRDWQERQNALAMQFNAQEAAKSRSWQEYMSN 75
+ N + +SR+ A N A SAQ A QR+WQ +QNA AMQFNA+EAAK+RSWQE+MSN
Sbjct 7 INNAANQVSRMQGIAQANNAWSAQQAQIQREWQVQQNAKAMQFNAEEAAKNRSWQEFMSN 66
Query 76 TAHQREIRDLKAAGLNPVLSAMggngaavtsgatasgvtsagaKGEVDTSANAALVQMLG 135
TAHQRE+RDL AAGLNPVLSAM GNGAAV SGATASGVTS+GAKG+ DTS + A+ +LG
Sbjct 67 TAHQREVRDLMAAGLNPVLSAMNGNGAAVGSGATASGVTSSGAKGDTDTSTSGAIANLLG 126
Query 136 SVLSAQTQLQTANVNARTQEAVADKYTAMEEIVANISRDATLGSAGIHagatryaadtsa 195
S++SA L++AN+NARTQEAVADKY AM +IVA I++ ATLGSAGIH A
Sbjct 127 SLVSASQALESANINARTQEAVADKYNAMSQIVAEINKSATLGSAGIH-----------A 175
Query 196 aasryfaDKNYEGTKYSSDKHYQ-----------GTMYSANKSYEGTKYSSDNS 238
AS+Y AD+ T YS+D+H +M+ + +S ++Y+SD S
Sbjct 176 GASKYAADRGAAATMYSADQHRAAAKYSADAAKLASMFGSIQSSSASRYASDQS 229
> Gokush_Human_gut_32_030_Microviridae_AG0186_putative.VP2
Length=289
Score = 191 bits (484), Expect = 3e-60, Method: Compositional matrix adjust.
Identities = 131/238 (55%), Positives = 164/238 (69%), Gaps = 11/238 (5%)
Query 10 VQSVPAVGNLDSALSRITRTASENTAKSAQMASEQRDWQERQNALAMQFNAQEAAKSRSW 69
V + PAV + + A N+A +A+ A QRDW E A M+FN+ EAAK+R W
Sbjct 15 VGAAPAVNRAADQIVSLKGVAQANSAFNAEQAKLQRDWTESMTARQMEFNSAEAAKNRQW 74
Query 70 QEYMSNTAHQREIRDLKAAGLNPVLSAMggngaavtsgatasgvtsagaKGEVDTSANAA 129
QE MSNTAHQRE+RDL AAGLNPVLSAM GNGAAV SGATAS +G+K + DT+A+ A
Sbjct 75 QEMMSNTAHQREVRDLMAAGLNPVLSAMNGNGAAVGSGATASASLGSGSKADADTAASGA 134
Query 130 LVQMLGSVLSAQTQLQTANVNARTQEAVADKYTAMEEIVANISRDATLGSAGIHagatry 189
+ +LGS+L AQT LQ+AN+NARTQEAVADKYTAME IVA I+ A + AGIHAGATR
Sbjct 135 IANLLGSILGAQTALQSANINARTQEAVADKYTAMEHIVAQIAAAAGIKQAGIHAGATRD 194
Query 190 aadtsaaasryfaDK-----------NYEGTKYSSDKHYQGTMYSANKSYEGTKYSSD 236
AA S++A+RY A + N T+YS+D+H GT Y A+KS + +KY+SD
Sbjct 195 AAAMSSSATRYAAGQAALASMFGSSVNSAATRYSADQHLSGTKYGADKSSDASKYASD 252
> Gokush_Human_gut_31_045_Microviridae_AG0115_putative.VP2
Length=289
Score = 191 bits (484), Expect = 3e-60, Method: Compositional matrix adjust.
Identities = 131/238 (55%), Positives = 164/238 (69%), Gaps = 11/238 (5%)
Query 10 VQSVPAVGNLDSALSRITRTASENTAKSAQMASEQRDWQERQNALAMQFNAQEAAKSRSW 69
V + PAV + + A N+A +A+ A QRDW E A M+FN+ EAAK+R W
Sbjct 15 VGAAPAVNRAADQIVSLKGVAQANSAFNAEQAKLQRDWTESMTARQMEFNSAEAAKNRQW 74
Query 70 QEYMSNTAHQREIRDLKAAGLNPVLSAMggngaavtsgatasgvtsagaKGEVDTSANAA 129
QE MSNTAHQRE+RDL AAGLNPVLSAM GNGAAV SGATAS +G+K + DT+A+ A
Sbjct 75 QEMMSNTAHQREVRDLMAAGLNPVLSAMNGNGAAVGSGATASASLGSGSKADADTAASGA 134
Query 130 LVQMLGSVLSAQTQLQTANVNARTQEAVADKYTAMEEIVANISRDATLGSAGIHagatry 189
+ +LGS+L AQT LQ+AN+NARTQEAVADKYTAME IVA I+ A + AGIHAGATR
Sbjct 135 IANLLGSILGAQTALQSANINARTQEAVADKYTAMEHIVAQIAAAAGIKQAGIHAGATRD 194
Query 190 aadtsaaasryfaDK-----------NYEGTKYSSDKHYQGTMYSANKSYEGTKYSSD 236
AA S++A+RY A + N T+YS+D+H GT Y A+KS + +KY+SD
Sbjct 195 AAAMSSSATRYAAGQAALASMFGSSVNSAATRYSADQHLSGTKYGADKSSDASKYASD 252
> Gokush_Human_gut_27_035_Microviridae_AG0164_putative.VP2
Length=289
Score = 191 bits (484), Expect = 3e-60, Method: Compositional matrix adjust.
Identities = 131/238 (55%), Positives = 164/238 (69%), Gaps = 11/238 (5%)
Query 10 VQSVPAVGNLDSALSRITRTASENTAKSAQMASEQRDWQERQNALAMQFNAQEAAKSRSW 69
V + PAV + + A N+A +A+ A QRDW E A M+FN+ EAAK+R W
Sbjct 15 VGAAPAVNRAADQIVSLKGVAQANSAFNAEQAKLQRDWTESMTARQMEFNSAEAAKNRQW 74
Query 70 QEYMSNTAHQREIRDLKAAGLNPVLSAMggngaavtsgatasgvtsagaKGEVDTSANAA 129
QE MSNTAHQRE+RDL AAGLNPVLSAM GNGAAV SGATAS +G+K + DT+A+ A
Sbjct 75 QEMMSNTAHQREVRDLMAAGLNPVLSAMNGNGAAVGSGATASASLGSGSKADADTAASGA 134
Query 130 LVQMLGSVLSAQTQLQTANVNARTQEAVADKYTAMEEIVANISRDATLGSAGIHagatry 189
+ +LGS+L AQT LQ+AN+NARTQEAVADKYTAME IVA I+ A + AGIHAGATR
Sbjct 135 IANLLGSILGAQTALQSANINARTQEAVADKYTAMEHIVAQIAAAAGIKQAGIHAGATRD 194
Query 190 aadtsaaasryfaDK-----------NYEGTKYSSDKHYQGTMYSANKSYEGTKYSSD 236
AA S++A+RY A + N T+YS+D+H GT Y A+KS + +KY+SD
Sbjct 195 AAAMSSSATRYAAGQAALASMFGSSVNSAATRYSADQHLSGTKYGADKSSDASKYASD 252
> Gokush_Human_feces_E_009_Microviridae_AG0283_putative.VP2
Length=294
Score = 151 bits (382), Expect = 4e-45, Method: Compositional matrix adjust.
Identities = 102/155 (66%), Positives = 120/155 (77%), Gaps = 0/155 (0%)
Query 17 GNLDSALSRITRTASENTAKSAQMASEQRDWQERQNALAMQFNAQEAAKSRSWQEYMSNT 76
G +++AL I A N A SA A + R WQE QN AM+FNA EAAK+R WQEYMSNT
Sbjct 30 GAINNALDSIAGIAKYNNAYSASQAQDLRSWQEEQNRKAMEFNAAEAAKNRDWQEYMSNT 89
Query 77 AHQREIRDLKAAGLNPVLSAMggngaavtsgatasgvtsagaKGEVDTSANAALVQMLGS 136
AHQREI DLKAAGLNPVLSA GGNGAAVTSGATASGVTS+GAKG+VDTS NAAL +LG+
Sbjct 90 AHQREIADLKAAGLNPVLSATGGNGAAVTSGATASGVTSSGAKGDVDTSVNAALASILGT 149
Query 137 VLSAQTQLQTANVNARTQEAVADKYTAMEEIVANI 171
+ + + L+ A+VNA+ AVA+KYTAM E+VA I
Sbjct 150 LWNNENALKIADVNAKNNLAVAEKYTAMNELVAQI 184
> Gokush_Human_feces_B_068_Microviridae_AG0332_putative.VP2
Length=346
Score = 118 bits (296), Expect = 2e-32, Method: Compositional matrix adjust.
Identities = 78/128 (61%), Positives = 97/128 (76%), Gaps = 0/128 (0%)
Query 55 AMQFNAQEAAKSRSWQEYMSNTAHQREIRDLKAAGLNPVLSAMggngaavtsgatasgvt 114
AM FN +EA K+R WQ+YMS+TAHQREI+DL+AAGLNPVLSAMGGNGA VTSGATASG
Sbjct 87 AMAFNREEAQKNRDWQQYMSDTAHQREIKDLQAAGLNPVLSAMGGNGAPVTSGATASGYA 146
Query 115 sagaKGEVDTSANAALVQMLGSVLSAQTQLQTANVNARTQEAVADKYTAMEEIVANISRD 174
S GAKG+ DTSA+ ALV +LGS++ +QTQL +A AVADKYT M++ V + +
Sbjct 147 SQGAKGDTDTSASGALVSLLGSLIQSQTQLANTATSANASLAVADKYTQMQKFVGELQAN 206
Query 175 ATLGSAGI 182
L ++ I
Sbjct 207 TQLTTSKI 214
> Gokush_Human_feces_A_029_Microviridae_AG0221_putative.VP2
Length=299
Score = 112 bits (279), Expect = 2e-30, Method: Compositional matrix adjust.
Identities = 78/145 (54%), Positives = 97/145 (67%), Gaps = 9/145 (6%)
Query 42 SEQRD----WQERQNALAMQFNAQEAAKSRSWQEYMSNTAHQREIRDLKAAGLNPVLSAM 97
+E RD W +Q AM+FN EA KSR WQEYMS+TAHQRE++DL AAGLNPVLSAM
Sbjct 44 TELRDYNNAWTAKQADKAMEFNRDEAEKSRKWQEYMSSTAHQREVKDLVAAGLNPVLSAM 103
Query 98 ggngaavtsgatasgvtsagaKGEVDTSANAALVQMLGSVLSAQTQLQTANVNARTQEAV 157
GG+GA VTSGATASG DTS ++ LVQ+ G++LS+QTQL ++A+T +V
Sbjct 104 GGSGAPVTSGATASGYAP-----SADTSLSSGLVQLFGALLSSQTQLANKALDAQTNLSV 158
Query 158 ADKYTAMEEIVANISRDATLGSAGI 182
ADKYT VA + L +A I
Sbjct 159 ADKYTETSRAVAQLQSQTQLTTANI 183
> Gokush_gi|393707865|ref|YP_004732987.1|_structural_protein_VP2_[Microviridae_phi-CA82]
Length=234
Score = 62.0 bits (149), Expect = 3e-13, Method: Compositional matrix adjust.
Identities = 64/235 (27%), Positives = 104/235 (44%), Gaps = 39/235 (17%)
Query 41 ASEQRDWQERQNALAMQFNAQEAAKSRSWQEYMSNTAHQREIRDLKAAGLNPVLSAMggn 100
A +Q W Q + QFNAQEA K+R WQE MSNTA QR+++D + AGLNP+ +
Sbjct 12 ADKQNKWNAEQTEKSNQFNAQEAQKNRDWQEQMSNTALQRKMQDAEKAGLNPIFA----- 66
Query 101 gaavtsgatasgvtsagaKGEVDTS---ANAALVQMLGSVLSAQTQLQTANVNARTQEAV 157
A+G TS A+A + +++A T A R Q +
Sbjct 67 ---------------LNAQGASTTSGATASADSSKPATDIINAMTSYTNAQEQNRMQREL 111
Query 158 ADKYTAMEEIVANISRDATLGSAGIHagatryaadtsaaasryfaDKNYEGTKYSSDKHY 217
A + ++ +A + A L A + A A SA ++Y +D+ Y
Sbjct 112 ALQENRVKLQIAQMQMQAELQRARLATNAQMATAYASA-----------SASRYGADRSY 160
Query 218 QGTMYSANKSYEGTKYSSDNSVR----NPSSAVGYAREIGKVAVNIFSDLFG-WD 267
+ YS++ SY+ + ++N++ NP+ + +E + N + F WD
Sbjct 161 NASRYSSDTSYKNVQSQNENNILTKGVNPTGVLYNNKEFQRRTNNAINKGFNLWD 215
> Gokush_Human_feces_A_019_Microviridae_AG0437_putative.VP2
Length=300
Score = 59.7 bits (143), Expect = 4e-12, Method: Compositional matrix adjust.
Identities = 24/40 (60%), Positives = 34/40 (85%), Gaps = 0/40 (0%)
Query 55 AMQFNAQEAAKSRSWQEYMSNTAHQREIRDLKAAGLNPVL 94
AM +N+ EAA +R WQEYMS+TA+QR + D++AAG+NP+L
Sbjct 133 AMAYNSAEAAHNREWQEYMSSTAYQRAVADMRAAGINPIL 172
> Gokush_Human_feces_E_017_Microviridae_AG0128_putative.VP2
Length=300
Score = 57.4 bits (137), Expect = 2e-11, Method: Compositional matrix adjust.
Identities = 23/40 (58%), Positives = 34/40 (85%), Gaps = 0/40 (0%)
Query 55 AMQFNAQEAAKSRSWQEYMSNTAHQREIRDLKAAGLNPVL 94
AM +N+ EAA +R WQE+MS+TA+QR + D++AAG+NP+L
Sbjct 133 AMAYNSAEAALNREWQEHMSSTAYQRAVADMRAAGINPIL 172
Lambda K H a alpha
0.309 0.121 0.329 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 22981276