bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: Microviridae_proteins.fasta
575 sequences; 147,441 total letters
Query= Contig-42_CDS_annotation_glimmer3.pl_2_2
Length=496
Score E
Sequences producing significant alignments: (Bits) Value
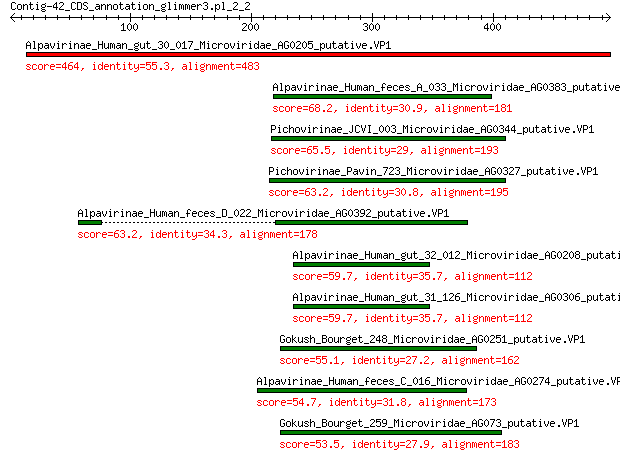
Alpavirinae_Human_gut_30_017_Microviridae_AG0205_putative.VP1 464 1e-158
Alpavirinae_Human_feces_A_033_Microviridae_AG0383_putative.VP1 68.2 5e-14
Pichovirinae_JCVI_003_Microviridae_AG0344_putative.VP1 65.5 3e-13
Pichovirinae_Pavin_723_Microviridae_AG0327_putative.VP1 63.2 2e-12
Alpavirinae_Human_feces_D_022_Microviridae_AG0392_putative.VP1 63.2 2e-12
Alpavirinae_Human_gut_32_012_Microviridae_AG0208_putative.VP1 59.7 2e-11
Alpavirinae_Human_gut_31_126_Microviridae_AG0306_putative.VP1 59.7 2e-11
Gokush_Bourget_248_Microviridae_AG0251_putative.VP1 55.1 7e-10
Alpavirinae_Human_feces_C_016_Microviridae_AG0274_putative.VP1 54.7 1e-09
Gokush_Bourget_259_Microviridae_AG073_putative.VP1 53.5 2e-09
> Alpavirinae_Human_gut_30_017_Microviridae_AG0205_putative.VP1
Length=647
Score = 464 bits (1194), Expect = 1e-158, Method: Compositional matrix adjust.
Identities = 267/516 (52%), Positives = 337/516 (65%), Gaps = 39/516 (8%)
Query 14 ASYVNSLFSWLRIGNKS--NIGGTPLASVSLPSNASMTQWS-NADTYLAYWDIVRNYYGY 70
A+ NSL SWLRI N N G PL S + S S NADTYL YWDIVRNYY Y
Sbjct 138 AALPNSLMSWLRIANSPIFNYSGNPLPSGGVLFKTSPKFLSVNADTYLGYWDIVRNYYSY 197
Query 71 SQWGLYSFAWPMANRLLY--SASAFALDPDNPAGSRFFTQCYGNLEFLDAFFESQFYPS- 127
S WG++SFA P R ++ S+ + + + + +F Q YGNLEFLD +FE+ FYP
Sbjct 198 SSWGVFSFAHPGTYRPVFYTGTSSSVTNVEYRSQASYFWQRYGNLEFLDHYFETMFYPRD 257
Query 128 -AVASTNN--TFNRGNLFYQIICSDL---DSTGSTGDGFPVKNIYPSTNLLGSTGIKSQI 181
VA+ + ++NR +LF +I+ SDL D+T ++ D + +YP +
Sbjct 258 RIVAADRDELSWNRSDLFVEILRSDLFNKDTTAASPDFTKMPQMYPEAMNFNVPAYPYDV 317
Query 182 QADTVNAAN---------SALAY-------FIVAHPMAVVPSNPDRFSRLIPTG-STSAV 224
Q V+ N S + + F+ AHPMAV PS+PDRFSRL+P G S S V
Sbjct 318 QEPKVDWNNGTGTDVNVPSKVFFAATLNVPFLAAHPMAVCPSSPDRFSRLMPPGDSNSDV 377
Query 225 SMSGVTTIPQLAIASRLQEYKDLLGAGGNRYSDWLDTFFASKIEHVDRPKLLFSASQTIN 284
+G+ TIPQLA+A+RLQEYKDL+GA G+RYSDWL TFFASKIEHVDRPKLLFS+S +N
Sbjct 378 DFTGLKTIPQLAVATRLQEYKDLIGASGSRYSDWLYTFFASKIEHVDRPKLLFSSSVMVN 437
Query 285 VQVIMNQAGPNNFSGLDISGPLGQQGGAIAFNEQLGRRQSYYFSEPGYLIDMLSIRPVYY 344
QV+MNQAG + F+G + + LGQ GG+IAFN LGR Q+YYF EPGY+ DML+IRPVY+
Sbjct 438 SQVVMNQAGQSGFAGGEAAA-LGQMGGSIAFNTVLGREQTYYFKEPGYIFDMLTIRPVYF 496
Query 345 WSFVKPDYLNYLGPDYFNPIYNDIGYQDVSSARIVFNGNAGATS----ASEPCFNEFRAS 400
W+ ++PDYL Y GPDYFNPIYNDIGYQDV R+ F + A S A EPC+NEFR+S
Sbjct 497 WTGIRPDYLEYRGPDYFNPIYNDIGYQDVPLWRLGFGWKSDAVSSVSVAKEPCYNEFRSS 556
Query 401 YDEVLGQLQGYPMPEVDGTSYIPLYAYWVQQRTSKLSDGSGSLPESHYYPILFTDMNQVN 460
YDEVLG LQ P+ +PL +YWVQQR L S L E +LFT++ VN
Sbjct 557 YDEVLGSLQSTLTPKAS----VPLQSYWVQQRDFYLIGLSSDLNEIS-PSMLFTNLATVN 611
Query 461 SPFASKVEDNFFVNMSYAVQKKSLVNKTFATRLSNR 496
+PF+S +EDNFFVNMSY V K+LVNK+FATRLS+R
Sbjct 612 NPFSSDMEDNFFVNMSYKVVVKNLVNKSFATRLSSR 647
> Alpavirinae_Human_feces_A_033_Microviridae_AG0383_putative.VP1
Length=590
Score = 68.2 bits (165), Expect = 5e-14, Method: Compositional matrix adjust.
Identities = 56/183 (31%), Positives = 91/183 (50%), Gaps = 16/183 (9%)
Query 218 TGSTSAVSMSGVTTIPQLAIASRLQEYKDLLGAGGNRYSDWLDTFFASKIEHVDR-PKLL 276
+ S S+VSM +T ASR+Q Y DL AGG R SD+ ++ F K++ + P L
Sbjct 284 SASGSSVSMRNIT------FASRMQRYMDLAFAGGGRNSDFYESQFDVKLDQDNTCPAFL 337
Query 277 FSASQTINVQVIMNQAGPNNFSGLDISGPLGQQGGAIAFNEQLGRRQSYYFSEPGYLIDM 336
S S +NV + G D S PLG G ++ + RR++Y+F++ GY +++
Sbjct 338 GSDSFDMNVNTLYQTTGFE-----DNSSPLGAFSGQLSGGTRF-RRRNYHFNDDGYFMEI 391
Query 337 LSIRP-VYYWSFVKPDYLNYLGPDYFNPIYNDIGYQDVSSARIVFNGNAGATSASEPCFN 395
SI P VYY S++ P + P ++I Q + ++ + G + A+ P +
Sbjct 392 TSIVPRVYYPSYINPTSRQISLGQQYAPALDNIAMQGLKASTVF--GEVQSLGATNPSYA 449
Query 396 EFR 398
E R
Sbjct 450 ENR 452
> Pichovirinae_JCVI_003_Microviridae_AG0344_putative.VP1
Length=505
Score = 65.5 bits (158), Expect = 3e-13, Method: Compositional matrix adjust.
Identities = 56/197 (28%), Positives = 89/197 (45%), Gaps = 15/197 (8%)
Query 217 PTGSTSAVSMSGVTTIPQLAIASRLQEYKDLLGAGGNRYSDWLDTFF--ASKIEHVDRPK 274
P GS S TTI L A +LQE+ +LL GG+RY + + TFF S + + RP+
Sbjct 255 PDGSLEVTS----TTITDLRRAFKLQEWLELLARGGSRYIEQIKTFFDVKSSDQRLQRPE 310
Query 275 LLFSASQTINVQVIMNQAGPNNFSGLDISGPLGQQGGAIAFNEQLGRRQSYYFSEPGYLI 334
+ Q + + I+N G +GL G + G A+ G YY E G++I
Sbjct 311 YITGTKQPVIISEILNTTGET--AGLP-QGNMSGHGVAVG----TGNVGKYYCEEHGFII 363
Query 335 DMLSIRP-VYYWSFVKPDYLNYLGPDYFNPIYNDIGYQDVSSARI-VFNGNAGATSASEP 392
+LS+ P Y + YL D++ P + IG Q + + I ++ T P
Sbjct 364 GILSVTPNTAYQQGIPKHYLRTDKYDFYWPQFAHIGEQPIVNNEIYAYDPTGDETFGYTP 423
Query 393 CFNEFRASYDEVLGQLQ 409
+ E++ V G+ +
Sbjct 424 RYAEYKYMPSRVAGEFR 440
> Pichovirinae_Pavin_723_Microviridae_AG0327_putative.VP1
Length=508
Score = 63.2 bits (152), Expect = 2e-12, Method: Compositional matrix adjust.
Identities = 60/218 (28%), Positives = 96/218 (44%), Gaps = 35/218 (16%)
Query 215 LIPTGSTSAVSMSGV--------------TTIPQLAIASRLQEYKDLLGAGGNRYSDWLD 260
LI G ++AV+ G TTI L A +LQE+ + GG RY + +
Sbjct 238 LIDIGGSNAVNADGTPTMIAKTSTIDIEPTTINDLRRAFKLQEWLEKNARGGTRYIENIL 297
Query 261 TFFA--SKIEHVDRPKLLFSASQTINVQVIMNQAGPNNFSGLDISGPLGQQGGAIAFNEQ 318
T F S + + RP+ + + V ++N G + GL P G G +
Sbjct 298 THFGVRSSDKRLQRPEYITGVKSPVVVSEVLNTTGQD--GGL----PQGNMAGH-GISVT 350
Query 319 LGRRQSYYFSEPGYLIDMLSIRPVYYW------SFVKPDYLNYLGPDYFNPIYNDIGYQD 372
G+ SYY E GY+I ++S+ P + +F+K D L DYF P + +IG Q+
Sbjct 351 SGKSGSYYCEEHGYIIGIMSVMPKTAYQQGIPRTFLKTDSL-----DYFWPTFANIGEQE 405
Query 373 VSSARI-VFNGNAGATSASEPCFNEFRASYDEVLGQLQ 409
V+ + + NA T P + E++ V G+ +
Sbjct 406 VAKQELYAYTANANDTFGYVPRYAEYKYMPSRVAGEFR 443
> Alpavirinae_Human_feces_D_022_Microviridae_AG0392_putative.VP1
Length=589
Score = 63.2 bits (152), Expect = 2e-12, Method: Compositional matrix adjust.
Identities = 51/161 (32%), Positives = 80/161 (50%), Gaps = 14/161 (9%)
Query 220 STSAVSMSGVTTIPQLAIASRLQEYKDLLGAGGNRYSDWLDTFFASKIEHVDR-PKLLFS 278
S S+VSM +T ASR+Q Y DL AGG R SD+ + F K+ + P L S
Sbjct 285 SGSSVSMRNIT------FASRMQRYMDLAFAGGGRNSDFYEAQFDVKLNQDNTCPAFLGS 338
Query 279 ASQTINVQVIMNQAGPNNFSGLDISGPLGQQGGAIAFNEQLGRRQSYYFSEPGYLIDMLS 338
S +NV + G D S PLG G ++ + RR++Y+F++ GY +++ S
Sbjct 339 DSFDMNVNTLYQTTGFE-----DNSSPLGAFSGQLSGGTRF-RRRNYHFNDDGYFMEITS 392
Query 339 IRP-VYYWSFVKPDYLNYLGPDYFNPIYNDIGYQDVSSARI 378
I P VYY S++ P + P ++I Q + ++ +
Sbjct 393 IVPRVYYPSYINPTSRQISLGQQYAPALDNIAMQGLKASTV 433
Score = 22.7 bits (47), Expect = 7.4, Method: Compositional matrix adjust.
Identities = 10/19 (53%), Positives = 12/19 (63%), Gaps = 0/19 (0%)
Query 57 YLAYWDIVRNYYGYSQWGL 75
Y+ Y DI NYY Q+GL
Sbjct 161 YIGYIDIYYNYYLNQQFGL 179
> Alpavirinae_Human_gut_32_012_Microviridae_AG0208_putative.VP1
Length=604
Score = 59.7 bits (143), Expect = 2e-11, Method: Compositional matrix adjust.
Identities = 40/113 (35%), Positives = 59/113 (52%), Gaps = 7/113 (6%)
Query 235 LAIASRLQEYKDLLGAGGNRYSDWLDTFF-ASKIEHVDRPKLLFSASQTINVQVIMNQAG 293
+A S +Q + DL AGG+RYSD++++ F S+++H P L S Q + VI G
Sbjct 300 IAAQSHIQRWLDLALAGGSRYSDYINSQFDVSRLKHTTSPLYLGSDRQYLGSNVIYQTTG 359
Query 294 PNNFSGLDISGPLGQQGGAIAFNEQLGRRQSYYFSEPGYLIDMLSIRPVYYWS 346
D + PLG G + E RR+SY+F E GY + M S+ P +S
Sbjct 360 AG-----DSASPLGAFAGQSSGGETF-RRRSYHFGENGYFVVMASLVPDVVYS 406
> Alpavirinae_Human_gut_31_126_Microviridae_AG0306_putative.VP1
Length=604
Score = 59.7 bits (143), Expect = 2e-11, Method: Compositional matrix adjust.
Identities = 40/113 (35%), Positives = 59/113 (52%), Gaps = 7/113 (6%)
Query 235 LAIASRLQEYKDLLGAGGNRYSDWLDTFF-ASKIEHVDRPKLLFSASQTINVQVIMNQAG 293
+A S +Q + DL AGG+RYSD++++ F S+++H P L S Q + VI G
Sbjct 300 IAAQSHIQRWLDLALAGGSRYSDYINSQFDVSRLKHTTSPLYLGSDRQYLGSNVIYQTTG 359
Query 294 PNNFSGLDISGPLGQQGGAIAFNEQLGRRQSYYFSEPGYLIDMLSIRPVYYWS 346
D + PLG G + E RR+SY+F E GY + M S+ P +S
Sbjct 360 AG-----DSASPLGAFAGQSSGGETF-RRRSYHFGENGYFVVMASLVPDVVYS 406
> Gokush_Bourget_248_Microviridae_AG0251_putative.VP1
Length=546
Score = 55.1 bits (131), Expect = 7e-10, Method: Compositional matrix adjust.
Identities = 44/165 (27%), Positives = 76/165 (46%), Gaps = 7/165 (4%)
Query 224 VSMSGVTTIPQLAIASRLQEYKDLLGAGGNRYSDWLDTFFA--SKIEHVDRPKLLFSASQ 281
+S++ TI QL + ++Q+ + GG RY++ + + F S + RP+ + S
Sbjct 286 LSVATAATINQLRQSFQIQKLLERDARGGTRYTEIIRSHFGVISPDARLQRPEYIGGGST 345
Query 282 TINVQVIMNQAGPNNFSGLDISGPLGQQGGAIAFNEQLGRRQSYYFSEPGYLIDMLSIRP 341
IN+ I +G N G L G A+A N +Y +E G +I M+S+R
Sbjct 346 NININPIAQTSGTNASGTTTPMGTLAAMGTALAHNHGF----TYSSTEHGVIIGMVSVRA 401
Query 342 -VYYWSFVKPDYLNYLGPDYFNPIYNDIGYQDVSSARIVFNGNAG 385
+ Y ++ + D++ P + +G Q V + I GNAG
Sbjct 402 DLTYQQGLQRMWSRSTRYDFYFPAFATLGEQAVLNKEIYVTGNAG 446
> Alpavirinae_Human_feces_C_016_Microviridae_AG0274_putative.VP1
Length=641
Score = 54.7 bits (130), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 55/193 (28%), Positives = 82/193 (42%), Gaps = 26/193 (13%)
Query 205 VPSNPDRFSRLIPTGSTSAVSMSGVTT------------IPQLAIASRLQEYKDLLGAGG 252
VP +PD F +I GS+ + + +P+L + +++Q + D L G
Sbjct 318 VPYSPDLFGNIIKQGSSPTAEIEVIKALDSNTDTGFSIAVPELRLKTKIQNWMDRLFISG 377
Query 253 NRYSDWLDTFFASK--IEHVDRPKLLFSASQTINVQVIMNQAGPNNFSGLDISGPLGQQG 310
R D T + +K +V++P L +IN + A + SG D + LGQ
Sbjct 378 GRVGDVFRTLWGTKSSAPYVNKPDFLGVWQASINPSNVRAMAN-GSASGEDAN--LGQLA 434
Query 311 GAI----AFNEQLGRRQSYYFSEPG--YLIDMLSIRPVYYWSFVKPDYLNYLGPDYFNPI 364
+ F++ G YY EPG LI ML P Y + PD + D FNP
Sbjct 435 ACVDRYCDFSDHSG--IDYYAKEPGTFMLITMLVPEPAYSQG-LHPDLASISFGDDFNPE 491
Query 365 YNDIGYQDVSSAR 377
N IG+Q V R
Sbjct 492 LNGIGFQLVPRHR 504
> Gokush_Bourget_259_Microviridae_AG073_putative.VP1
Length=540
Score = 53.5 bits (127), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 51/191 (27%), Positives = 88/191 (46%), Gaps = 14/191 (7%)
Query 224 VSMSGVTTIPQLAIASRLQEYKDLLGAGGNRYSDWLDTFF--ASKIEHVDRPKLLFSASQ 281
+S + TI QL + ++Q+ + GG RY++ + + F AS + RP+ L S
Sbjct 279 LSAATAATINQLRQSFQIQKLLERDARGGTRYTEIIRSHFGVASPDARLQRPEYLGGGST 338
Query 282 TINVQVIMNQAGPNNFSGLDISGPLGQQGGAIAFNEQLGRRQSYY--FSEPGYLIDMLSI 339
IN+ I N+ +GL SG G AF LG+ + F E GY+I ++S+
Sbjct 339 NINISPIAQ----NSATGL--SGSTTPIGNLAAFGTFLGKEHGFTQSFVEHGYVIGLVSV 392
Query 340 RP-VYYWSFVKPDYLNYLGPDYFNPIYNDIGYQDVSSARIVFNGNAGATSAS---EPCFN 395
R + Y ++ + DY+ P + +G Q V + I GN ++ + +
Sbjct 393 RADLTYQQGLRRHWSRSTRYDYYFPAFATLGEQSVLNKEIYVTGNTTQDNSVFGYQERWA 452
Query 396 EFRASYDEVLG 406
E+R + E+ G
Sbjct 453 EYRYNPSEITG 463
Lambda K H a alpha
0.318 0.133 0.403 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 44961384