bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: All non-redundant GenBank CDS translations+PDB+SwissProt+PIR+PRF
excluding environmental samples from WGS projects
49,011,213 sequences; 17,563,301,199 total letters
Query= Contig-10_CDS_annotation_glimmer3.pl_2_6
Length=111
Score E
Sequences producing significant alignments: (Bits) Value
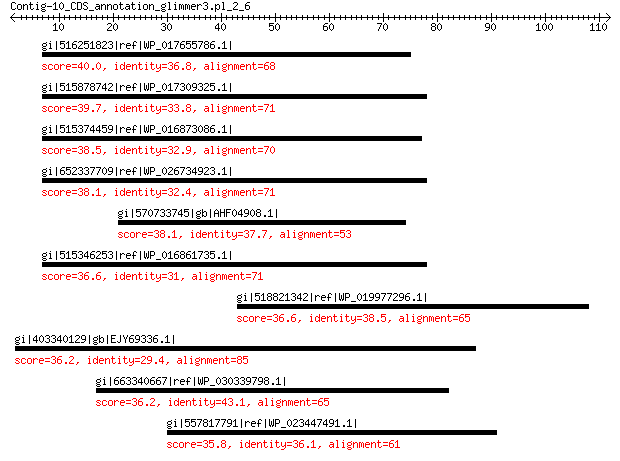
gi|516251823|ref|WP_017655786.1| hypothetical protein 40.0 0.097
gi|515878742|ref|WP_017309325.1| hypothetical protein 39.7 0.15
gi|515374459|ref|WP_016873086.1| hypothetical protein 38.5 0.30
gi|652337709|ref|WP_026734923.1| hypothetical protein 38.1 0.52
gi|570733745|gb|AHF04908.1| cobalamin-binding protein 38.1 0.65
gi|515346253|ref|WP_016861735.1| hypothetical protein 36.6 1.7
gi|518821342|ref|WP_019977296.1| sulfatase 36.6 1.8
gi|403340129|gb|EJY69336.1| hypothetical protein OXYTRI_10043 36.2 2.3
gi|663340667|ref|WP_030339798.1| PAS/PAC sensor protein 36.2 2.4
gi|557817791|ref|WP_023447491.1| hypothetical protein 35.8 4.1
>gi|516251823|ref|WP_017655786.1| hypothetical protein [Microchaete sp. PCC 7126]
Length=231
Score = 40.0 bits (92), Expect = 0.097, Method: Compositional matrix adjust.
Identities = 25/76 (33%), Positives = 38/76 (50%), Gaps = 11/76 (14%)
Query 7 FPKPPETNYEFQDGESIENKVRRITENNEPIT-----DGAPIIYT---NRDDGVLPAYNI 58
F P E+N+ IEN V R E +E I DG+P+I + N+ DGV+ + +
Sbjct 5 FFCPEESNFY---ANCIENLVLRNCEQSESIVEFGSGDGSPVINSLLRNKFDGVVHGFEL 61
Query 59 RTDRWEVAQAAMDAVN 74
T W+ A + +D N
Sbjct 62 NTSAWKTANSTIDEYN 77
>gi|515878742|ref|WP_017309325.1| hypothetical protein [Fischerella sp. PCC 9339]
Length=231
Score = 39.7 bits (91), Expect = 0.15, Method: Compositional matrix adjust.
Identities = 24/79 (30%), Positives = 40/79 (51%), Gaps = 11/79 (14%)
Query 7 FPKPPETNYEFQDGESIENKVRRITENNEPIT-----DGAPIIYT---NRDDGVLPAYNI 58
F P E+N+ +EN R + +E I DG+P+I + N+ DG++ + +
Sbjct 5 FFCPEESNFY---SNCLENFALRNCKKSECIVEFGSGDGSPVINSLLRNKFDGMIQGFEL 61
Query 59 RTDRWEVAQAAMDAVNQAN 77
T W+VA + +D N AN
Sbjct 62 NTSAWKVANSTIDEYNLAN 80
>gi|515374459|ref|WP_016873086.1| hypothetical protein [Chlorogloeopsis fritschii]
Length=233
Score = 38.5 bits (88), Expect = 0.30, Method: Compositional matrix adjust.
Identities = 23/78 (29%), Positives = 37/78 (47%), Gaps = 11/78 (14%)
Query 7 FPKPPETNYEFQDGESIENKVRRITENNEPIT-----DGAPII---YTNRDDGVLPAYNI 58
F P E+N+ +EN R + +E I DG P+I NR DG++ + +
Sbjct 5 FFCPEESNFY---SNCLENLALRNCQKSECIVEFGSGDGTPVINSLLRNRFDGIIHGFEL 61
Query 59 RTDRWEVAQAAMDAVNQA 76
T W+VA + +D N +
Sbjct 62 NTSAWKVANSTIDEYNLS 79
>gi|652337709|ref|WP_026734923.1| hypothetical protein [Fischerella sp. PCC 9605]
Length=234
Score = 38.1 bits (87), Expect = 0.52, Method: Compositional matrix adjust.
Identities = 23/79 (29%), Positives = 38/79 (48%), Gaps = 11/79 (14%)
Query 7 FPKPPETNYEFQDGESIENKVRRITENNEPIT-----DGAPII---YTNRDDGVLPAYNI 58
F P E+N+ +EN R + +E I DG+P+I NR DGV+ + +
Sbjct 5 FFCPEESNFY---SNCLENFALRNCKKSECIVEFGSGDGSPVINSLLRNRFDGVIHGFEL 61
Query 59 RTDRWEVAQAAMDAVNQAN 77
T W+ A + +D N ++
Sbjct 62 NTSAWKAANSTIDEYNLSH 80
>gi|570733745|gb|AHF04908.1| cobalamin-binding protein [Marichromatium purpuratum 984]
Length=530
Score = 38.1 bits (87), Expect = 0.65, Method: Composition-based stats.
Identities = 20/57 (35%), Positives = 34/57 (60%), Gaps = 4/57 (7%)
Query 21 ESIENKVRRI---TENNEPITDGAPIIYTNRDDGVLPAYNIRTDR-WEVAQAAMDAV 73
+S +++R+ TE+NE + GA +++ +RD + P Y++ TD WE AMD V
Sbjct 334 QSGSERIKRLYDRTESNERVVAGAELLHRHRDRLLPPDYHVITDNPWETEADAMDTV 390
>gi|515346253|ref|WP_016861735.1| hypothetical protein [Fischerella muscicola]
Length=231
Score = 36.6 bits (83), Expect = 1.7, Method: Compositional matrix adjust.
Identities = 22/79 (28%), Positives = 40/79 (51%), Gaps = 11/79 (14%)
Query 7 FPKPPETNYEFQDGESIENKVRRITENNEPIT-----DGAPIIYT---NRDDGVLPAYNI 58
F P E+++ +EN R + +E I DG+P+I + N+ DG++ + +
Sbjct 5 FFCPEESHFY---SNCLENFALRNCKKSECIVEFGSGDGSPVINSLLRNKFDGMIQGFEL 61
Query 59 RTDRWEVAQAAMDAVNQAN 77
T W+VA + +D N A+
Sbjct 62 NTSAWKVANSTIDEYNLAH 80
>gi|518821342|ref|WP_019977296.1| sulfatase [alpha proteobacterium SCGC AAA300-J04]
Length=504
Score = 36.6 bits (83), Expect = 1.8, Method: Compositional matrix adjust.
Identities = 25/69 (36%), Positives = 34/69 (49%), Gaps = 4/69 (6%)
Query 43 IIYTNRDDGVLPAYNIRTD--RWEVAQAAMDAVNQANLAKSKNYGKIEQQEQNALESK-- 98
IIY D LP N + WE+ D + NLAK +NYG+I Q + L K
Sbjct 435 IIYWYNDGYDLPGTNHGGEDKEWELFDCQNDPLELFNLAKDQNYGEIFSQMKLLLTEKML 494
Query 99 EIGDTPSQQ 107
+IGD P+ +
Sbjct 495 KIGDIPAHE 503
>gi|403340129|gb|EJY69336.1| hypothetical protein OXYTRI_10043 [Oxytricha trifallax]
Length=422
Score = 36.2 bits (82), Expect = 2.3, Method: Compositional matrix adjust.
Identities = 25/89 (28%), Positives = 39/89 (44%), Gaps = 4/89 (4%)
Query 2 IKPKFFPKPPETNYE----FQDGESIENKVRRITENNEPITDGAPIIYTNRDDGVLPAYN 57
+ P+F+ K +TN E + S +N ++ + N P G P+I + ++
Sbjct 201 VNPQFYDKETQTNMEDLDQLRQSVSTDNTLKEKRQMNSPQQTGRPVIVGSSQPKLVGLKR 260
Query 58 IRTDRWEVAQAAMDAVNQANLAKSKNYGK 86
DR E AQ DA + NL KN K
Sbjct 261 TILDRSESAQQNCDAKSDGNLKMVKNLLK 289
>gi|663340667|ref|WP_030339798.1| PAS/PAC sensor protein [Streptomyces sp. NRRL S-1022]
Length=690
Score = 36.2 bits (82), Expect = 2.4, Method: Composition-based stats.
Identities = 28/86 (33%), Positives = 37/86 (43%), Gaps = 21/86 (24%)
Query 17 FQDGESIENKVRRITENNEPITD----GAPIIYTNRDD-----------------GVLPA 55
F DGE+IE ++RR+ E EP+TD G P RD GV +
Sbjct 193 FLDGEAIEGRLRRVLETGEPVTDRYIVGRPASDPQRDHAWSVSYHRLEDAAGRVLGVATS 252
Query 56 YNIRTDRWEVAQAAMDAVNQANLAKS 81
T+R E A AA A N+ + S
Sbjct 253 VIDITERHEAAIAATQARNRLAMIAS 278
>gi|557817791|ref|WP_023447491.1| hypothetical protein [Asticcacaulis benevestitus]
gi|557339739|gb|ESQ79469.1| hypothetical protein ABENE_22670 [Asticcacaulis benevestitus
DSM 16100 = ATCC BAA-896]
Length=1077
Score = 35.8 bits (81), Expect = 4.1, Method: Compositional matrix adjust.
Identities = 22/66 (33%), Positives = 36/66 (55%), Gaps = 5/66 (8%)
Query 30 ITENNEPITDGAPIIYT--NRDDGVLPAYNIRTD---RWEVAQAAMDAVNQANLAKSKNY 84
++ N+ +GA II T + VLP++N++ D W V AA A+++ +L KNY
Sbjct 731 LSANDLAFNNGAYIIRTAKTKHTNVLPSFNLKIDLNDEWLVRFAASKAMSRPDLGYLKNY 790
Query 85 GKIEQQ 90
I +Q
Sbjct 791 SAITRQ 796
Lambda K H a alpha
0.308 0.128 0.363 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 438125291904