bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: All non-redundant GenBank CDS translations+PDB+SwissProt+PIR+PRF
excluding environmental samples from WGS projects
49,011,213 sequences; 17,563,301,199 total letters
Query= Contig-12_CDS_annotation_glimmer3.pl_2_1
Length=365
Score E
Sequences producing significant alignments: (Bits) Value
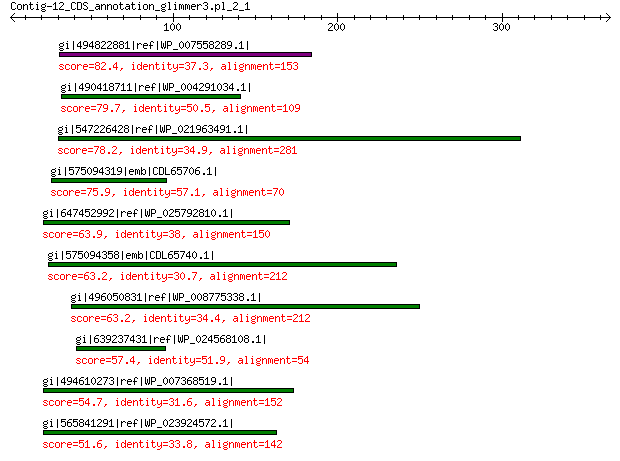
gi|494822881|ref|WP_007558289.1| hypothetical protein 82.4 2e-14
gi|490418711|ref|WP_004291034.1| hypothetical protein 79.7 2e-13
gi|547226428|ref|WP_021963491.1| putative uncharacterized protein 78.2 1e-12
gi|575094319|emb|CDL65706.1| unnamed protein product 75.9 7e-12
gi|647452992|ref|WP_025792810.1| hypothetical protein 63.9 5e-08
gi|575094358|emb|CDL65740.1| unnamed protein product 63.2 7e-08
gi|496050831|ref|WP_008775338.1| predicted protein 63.2 8e-08
gi|639237431|ref|WP_024568108.1| hypothetical protein 57.4 4e-06
gi|494610273|ref|WP_007368519.1| hypothetical protein 54.7 4e-05
gi|565841291|ref|WP_023924572.1| hypothetical protein 51.6 5e-04
>gi|494822881|ref|WP_007558289.1| hypothetical protein [Bacteroides plebeius]
gi|198272097|gb|EDY96366.1| hypothetical protein BACPLE_00802 [Bacteroides plebeius DSM 17135]
Length=344
Score = 82.4 bits (202), Expect = 2e-14, Method: Compositional matrix adjust.
Identities = 57/157 (36%), Positives = 85/157 (54%), Gaps = 8/157 (5%)
Query 31 TNQMNYKINQMNNQFNERMAIQQRNWQENMWNKENAYNTASAQRQRLEEAGLNPYLMMNG 90
TNQ N +I QM+N++N +Q + +MWN EN YN+AS+QR+RLEEAGLNPY+MM+G
Sbjct 43 TNQANIQIAQMSNEYNREQLERQIEQEWDMWNAENEYNSASSQRKRLEEAGLNPYMMMDG 102
Query 91 GSAG----VaqsagngataassgnaVMQPFQADYSGIGSSIGNIFQYDLMQSEKSQLQGA 146
GSAG + A A A MQP AD SG+ G ++ + ++G
Sbjct 103 GSAGSASSMTSPAAQPAVVPQMQGATMQP--ADMSGLSGLRGIASEFIATLKAQEDIRGQ 160
Query 147 RQLSDAKAMETLSNIDWGNLTAETRNYLRSTGLARAQ 183
+ +++ + +E D L A+ +G R+Q
Sbjct 161 QLINEGQEIENQYKAD--KLLADLEKTRTESGFVRSQ 195
>gi|490418711|ref|WP_004291034.1| hypothetical protein [Bacteroides eggerthii]
gi|217986638|gb|EEC52972.1| hypothetical protein BACEGG_02723 [Bacteroides eggerthii DSM
20697]
Length=368
Score = 79.7 bits (195), Expect = 2e-13, Method: Compositional matrix adjust.
Identities = 55/151 (36%), Positives = 70/151 (46%), Gaps = 42/151 (28%)
Query 32 NQMNYKINQMNNQFNERMAIQQ---------------------------------RNWQE 58
NQ N +I QMNN FNE+M +Q + +Q
Sbjct 31 NQANKEIAQMNNAFNEKMFDKQIAYNKEMYQTQLGDQWKFYDDQKANAWKLYEDNKAYQT 90
Query 59 NMWNKENAYNTASAQRQRLEEAGLNPYLMMNGGSAGVaqsagngataassg--------- 109
MWNK+N YN SAQR RLE AGLNPY+MMNGGSAGVA S +A S
Sbjct 91 EMWNKQNEYNDPSAQRARLEAAGLNPYMMMNGGSAGVAGSVSGTQGSAPSAGSPSAQGVQ 150
Query 110 naVMQPFQADYSGIGSSIGNIFQYDLMQSEK 140
P+ ADYSG+ +G+ + S++
Sbjct 151 PPTATPYSADYSGVMQGLGHAIDTIMTGSQR 181
>gi|547226428|ref|WP_021963491.1| putative uncharacterized protein [Prevotella sp. CAG:1185]
gi|524103380|emb|CCY83991.1| putative uncharacterized protein [Prevotella sp. CAG:1185]
Length=416
Score = 78.2 bits (191), Expect = 1e-12, Method: Compositional matrix adjust.
Identities = 98/348 (28%), Positives = 161/348 (46%), Gaps = 78/348 (22%)
Query 30 ETNQMNYKINQMNNQFNERMAIQQRNWQENMWNKE------------------------- 64
+TN+ N +I QMNN++NERM +Q + ++M+N++
Sbjct 28 DTNKTNLQIAQMNNEYNERMFNKQLEYNQDMFNQQVEYDQKKMEQQNNFNARMQNEAIGA 87
Query 65 -NAYNTASAQRQRLEEAGLNPYLMMNGGSAGVaqsagngat-----------aassgnaV 112
YN+A AQR RLE AGLNPYLMM+GG+AG + + ++ +AV
Sbjct 88 QQVYNSAKAQRARLEAAGLNPYLMMSGGNAGAVSAVSGSSGSGGSPSPMGVNPPTASSAV 147
Query 113 MQPFQADYSGIGSSIGNIFQYDLMQSEKS-----------QLQGARQLSDAKAMETL--- 158
MQ F+ D+SG+ I + +Q++K Q G + + KA + L
Sbjct 148 MQAFRPDFSGVTGIIQTLLD---IQAQKGVRDAQAFSLGEQASGFKIENKYKAEKLLWDI 204
Query 159 --SNIDWGNLTAETRNYLRSTGLARAQLGY----VKEQQEVDNMAMTGLIMRAQRSGMLL 212
S D+ NL ++ L + AR Q + K Q+E +N TG ++RAQ + L
Sbjct 205 YNSKADY-NLK-NSQESLNNMSFARLQAMFSSDVSKAQREAENAQFTGELIRAQTACQQL 262
Query 213 DNEAKGILN----KYLDQEQQLDLNVKAADYYQRMSAGYLSYaetkkaladealaaartr 268
+G+L KY DQ+ +L + +A Y ++AG S A+ ++A+ + +
Sbjct 263 ----QGLLGAKELKYYDQKVLQELAIMSAQQYSLVAAGKASEAQARQAIENALNLVEQRE 318
Query 269 GQKISNEVASRIAESQIAANIAANQSSEAY------HNEELKLGLSQD 310
G K+ N V + A + I A N + +Y HN+ L+ + +D
Sbjct 319 GIKVDNYVKQKTANALI--KTARNNCNTSYWNSKTAHNQSLRPSVFED 364
>gi|575094319|emb|CDL65706.1| unnamed protein product [uncultured bacterium]
Length=396
Score = 75.9 bits (185), Expect = 7e-12, Method: Compositional matrix adjust.
Identities = 40/70 (57%), Positives = 45/70 (64%), Gaps = 11/70 (16%)
Query 26 QNVRETNQMNYKINQMNNQFNERMAIQQRNWQENMWNKENAYNTASAQRQRLEEAGLNPY 85
Q VRETNQ N +I NN+FNERM WN +N YN QR RLE AGLNPY
Sbjct 54 QAVRETNQANREIADQNNKFNERM-----------WNLQNEYNRPDMQRARLEAAGLNPY 102
Query 86 LMMNGGSAGV 95
LMM+GGSAG+
Sbjct 103 LMMDGGSAGI 112
>gi|647452992|ref|WP_025792810.1| hypothetical protein [Prevotella histicola]
Length=424
Score = 63.9 bits (154), Expect = 5e-08, Method: Compositional matrix adjust.
Identities = 57/150 (38%), Positives = 77/150 (51%), Gaps = 13/150 (9%)
Query 21 NSQNRQNVRETNQMNYKINQMNNQFNERMAIQQRNWQENMWNKENAYNTASAQRQRLEEA 80
N N Q RETNQMNY++ Q +N FNE+M +++ NAYNT SAQ QR EA
Sbjct 30 NQTNLQIARETNQMNYQLFQESNAFNEKM-----------YHEANAYNTPSAQMQRYAEA 78
Query 81 GLNPYLMMNGGSAGVaqsagngataassgnaVMQPFQADYSGIGSSIGNIFQYDLMQSEK 140
G+NPY+ G SA A A A MQP +G+ S+ I QY Q+E
Sbjct 79 GINPYIAAGNVQTGNTTSALQSAPAPQMHAAQMQPDVNMANGMMSAGSIINQY--AQNEL 136
Query 141 SQLQGARQLSDAKAMETLSNIDWGNLTAET 170
+ Q + S+A ++ L+ GN+ A T
Sbjct 137 ALAQARKTNSEASWIDRLNTAYVGNMNAHT 166
>gi|575094358|emb|CDL65740.1| unnamed protein product [uncultured bacterium]
Length=328
Score = 63.2 bits (152), Expect = 7e-08, Method: Compositional matrix adjust.
Identities = 65/217 (30%), Positives = 102/217 (47%), Gaps = 40/217 (18%)
Query 24 NRQNVRETNQMNYKINQMNNQFNERMAIQQRNWQENMWNKENAYNTASAQRQRLEEAGLN 83
N+ V TN N +I QMNN+++ERM +Q + MW K YN+ + Q+ +AG+N
Sbjct 19 NKNAVDNTNSTNMQIAQMNNEWSERMMEKQMAYNTEMWEKVADYNSLPNKMQQARDAGVN 78
Query 84 PYLMMNGGSAGVaqsagngataassgnaV-MQPFQADYSGIGSSIGNIFQYDLMQSEKSQ 142
PY+ ++G + G + + + S + V QP Q D+S + +SI I DL Q K+Q
Sbjct 79 PYMALSGNAFGSISAPSANSVSLPSPSQVQAQPAQYDFSSVSNSI--IAGMDLFQ--KAQ 134
Query 143 LQGARQLSDAKAMETLSNIDWGNLTAETRNYLRSTGLARAQLGYVKEQQEVDNMAMTGLI 202
L ++Q SNID ST R + Y AM +
Sbjct 135 LMKSQQ----------SNID------------ASTDQLRIENKY---------HAMKLVS 163
Query 203 MRAQRSGMLLDNEAKG----ILNKYLDQEQQLDLNVK 235
A++ D++AK I+N+Y +Q + DL +K
Sbjct 164 EIAEKMANTKDSQAKAVYQQIINEYAEQGIKTDLEIK 200
>gi|496050831|ref|WP_008775338.1| predicted protein [Bacteroides sp. 2_2_4]
gi|229448895|gb|EEO54686.1| hypothetical protein BSCG_01611 [Bacteroides sp. 2_2_4]
Length=381
Score = 63.2 bits (152), Expect = 8e-08, Method: Compositional matrix adjust.
Identities = 73/246 (30%), Positives = 116/246 (47%), Gaps = 38/246 (15%)
Query 38 INQMNNQFNERMAIQQRNWQE----------------------NMWNKENAYNTASAQRQ 75
I QMNN+FNERM +Q ++ +M+N N YN+ASAQR+
Sbjct 41 IAQMNNEFNERMLQKQMDYNTLAYDQQVSDQWSFYNDAKQNAWDMFNATNEYNSASAQRE 100
Query 76 RLEEAGLNPYLMMNGGSAGVaqsagngataassgnaVM----QPFQADYSGIGSSIGN-I 130
R E AGLNPY+MMN GSAG A + + A + + P+ ADYSGI +G I
Sbjct 101 RYEAAGLNPYVMMNTGSAGTAAATSATSATAPTKQGITPPTASPYSADYSGIMQGLGQAI 160
Query 131 FQYDLMQSEKSQLQGARQLS---DAKAMETLSNIDWGNLTAETRNYLRSTGLARAQLGY- 186
Q + + + L KA E ++ I N+ A+T + + +A +L Y
Sbjct 161 DQLSSIPDKAKTIAETGNLKIEGKYKAAEAIARI--ANIKADTHS--KKEQVALNKLMYS 216
Query 187 VKEQQEVDNMAMTGLIMRAQRSGMLLDNEAKGILNK---YLDQEQQLDLNVKAADYYQRM 243
+++ MA+ + R+ N I +K ++D Q+++L KAA+ ++
Sbjct 217 IQKDLASSTMAVNSQNIANMRAEEKFKNIQTLIADKQLSFMDATQKMELAEKAANIQLKL 276
Query 244 SAGYLS 249
+ G L+
Sbjct 277 AQGALT 282
>gi|639237431|ref|WP_024568108.1| hypothetical protein [Elizabethkingia anophelis]
Length=287
Score = 57.4 bits (137), Expect = 4e-06, Method: Compositional matrix adjust.
Identities = 28/54 (52%), Positives = 37/54 (69%), Gaps = 0/54 (0%)
Query 41 MNNQFNERMAIQQRNWQENMWNKENAYNTASAQRQRLEEAGLNPYLMMNGGSAG 94
MNN N+++A + R + +MWN+ N YNT AQ QRL+EAGLNP LM G+ G
Sbjct 18 MNNSSNKKIARENRAFALDMWNRNNEYNTPLAQMQRLKEAGLNPNLMYGQGTTG 71
>gi|494610273|ref|WP_007368519.1| hypothetical protein [Prevotella multiformis]
gi|324988545|gb|EGC20508.1| hypothetical protein HMPREF9141_0987 [Prevotella multiformis
DSM 16608]
Length=437
Score = 54.7 bits (130), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 48/158 (30%), Positives = 74/158 (47%), Gaps = 21/158 (13%)
Query 21 NSQNRQNVRETNQMNYKINQMNNQFNERMAIQQRNWQENMWNKENAYNTASAQRQRLEEA 80
N N Q RE NQ Y++ Q N FNERM WN+ N YN+ +AQ QR +A
Sbjct 97 NETNLQIAREANQNQYQMFQEQNAFNERM-----------WNQMNQYNSPAAQMQRYTDA 145
Query 81 GLNPYLMMNGGSAGVaqsagngataassgnaVMQPFQADYSGIGSSIGNIFQ------YD 134
G+NPY+ G+ + +A + V Q A +G+G ++ N F
Sbjct 146 GINPYIA--AGNVQTGNAQSALQSAPAPQQHVAQVMPA--TGMGDAVQNSFAQIGNVISQ 201
Query 135 LMQSEKSQLQGARQLSDAKAMETLSNIDWGNLTAETRN 172
Q++ + Q + ++A ++ L++ G L AET N
Sbjct 202 FAQNQLALAQAKKTDAEASWIDRLNSAQMGKLGAETLN 239
>gi|565841291|ref|WP_023924572.1| hypothetical protein [Prevotella nigrescens]
gi|564729909|gb|ETD29853.1| hypothetical protein HMPREF1173_00035 [Prevotella nigrescens
CC14M]
Length=396
Score = 51.6 bits (122), Expect = 5e-04, Method: Compositional matrix adjust.
Identities = 48/146 (33%), Positives = 73/146 (50%), Gaps = 15/146 (10%)
Query 21 NSQNRQNVRETNQMNYKINQMNNQFNERMAIQQRNWQENMWNKENAYNTASAQRQRLEEA 80
NS N + RETN N+++ Q N+FN++M +K+N Y QR+R E+A
Sbjct 31 NSTNLRIARETNAANFQMMQYQNEFNQKM-----------LDKQNEYALPINQRKRFEDA 79
Query 81 GLNPYLMMNGGSAGVaqsagngataassgnaVMQPFQADYSGIGSSIGN-IFQY-DLMQS 138
G+NPY ++ S+G Q A A + A +QP A + S+ + + Y LMQ+
Sbjct 80 GINPYFALSQISSGTPQGALQSAQGHPAVAAQVQPVTAFGDALRDSVSHGVNTYGQLMQA 139
Query 139 EKSQLQGARQLSD--AKAMETLSNID 162
+ +Q Q Q + KA LS ID
Sbjct 140 KYTQQQAEGQSLENRFKAATLLSRID 165
Lambda K H a alpha
0.312 0.126 0.352 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 2216296179792