bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: All non-redundant GenBank CDS translations+PDB+SwissProt+PIR+PRF
excluding environmental samples from WGS projects
49,011,213 sequences; 17,563,301,199 total letters
Query= Contig-13_CDS_annotation_glimmer3.pl_2_3
Length=579
Score E
Sequences producing significant alignments: (Bits) Value
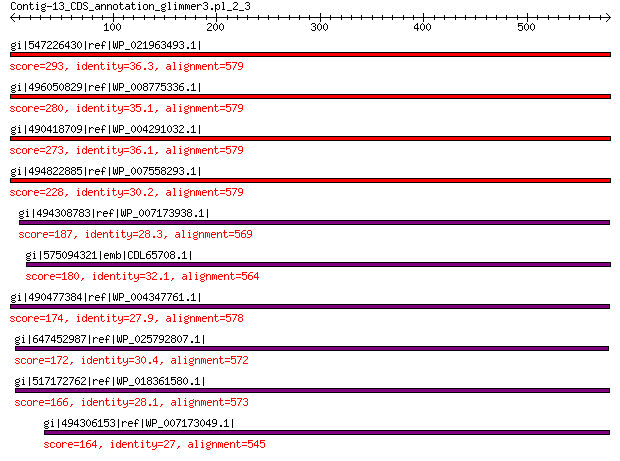
gi|547226430|ref|WP_021963493.1| putative uncharacterized protein 293 3e-87
gi|496050829|ref|WP_008775336.1| hypothetical protein 280 3e-82
gi|490418709|ref|WP_004291032.1| hypothetical protein 273 1e-79
gi|494822885|ref|WP_007558293.1| hypothetical protein 228 1e-62
gi|494308783|ref|WP_007173938.1| hypothetical protein 187 3e-48
gi|575094321|emb|CDL65708.1| unnamed protein product 180 1e-45
gi|490477384|ref|WP_004347761.1| capsid protein 174 1e-43
gi|647452987|ref|WP_025792807.1| hypothetical protein 172 3e-43
gi|517172762|ref|WP_018361580.1| hypothetical protein 166 6e-41
gi|494306153|ref|WP_007173049.1| hypothetical protein 164 8e-41
>gi|547226430|ref|WP_021963493.1| putative uncharacterized protein [Prevotella sp. CAG:1185]
gi|524103382|emb|CCY83994.1| putative uncharacterized protein [Prevotella sp. CAG:1185]
Length=573
Score = 293 bits (750), Expect = 3e-87, Method: Compositional matrix adjust.
Identities = 210/614 (34%), Positives = 296/614 (48%), Gaps = 76/614 (12%)
Query 1 MSDFNPLNRARISTHRSSFDLSSKKLFTAKVGEILPCYWQIAIPGNKYRISSDWFTRTVP 60
MS L + S R+ FDLS K FTAKVGE+LP + PG+K+ I FTRT P
Sbjct 1 MSSVMSLTALKNSVKRNGFDLSFKNAFTAKVGELLPIMCKEVYPGDKFNIRGQAFTRTQP 60
Query 61 VNTAAYTRIKEYYDFYAVPLRLISRALPQAFTQMTD--YItsaasstansstltsVPFVS 118
VN+AAY+R++EYYDFY VP RL+ P FT M D + SS S F
Sbjct 61 VNSAAYSRLREYYDFYFVPYRLLWNMAPTFFTNMPDPHHAADLVSSVNLSQRHPWFTFFD 120
Query 119 QTLFNAFFQTANAGDQPNTRDDAGLPIVYGSCKLLDMLGYGSMIASNNPSKAAITKKYLG 178
+ + + + ++ G V S KLL+ L YG K Y
Sbjct 121 IMEYLGNLNSLSGAYEKYQKNFFGFSRVELSVKLLNYLNYG------------FGKDYES 168
Query 179 VDSLNDADNPLVYQTSQTVNALPFLAYQKIYYDFFSNSQWEKHKAYAYNVDYWSGAGN-- 236
V +D+D+ ++ + P LAYQKI D+F + QW+ Y YN+DY G +
Sbjct 169 VKVPSDSDDIVL-------SPFPLLAYQKICEDYFRDDQWQSAAPYRYNLDYLYGKSSGF 221
Query 237 ----IGLVTD------MVQLRYANYPKDYFMGMLPSSQYGSVAVLPsissssdsrsLLYY 286
D M L Y N+ KDYF GMLP +QYG V+V
Sbjct 222 HIPMSSFTNDAFKNPTMFDLNYCNFQKDYFTGMLPRAQYGDVSVA--------------- 266
Query 287 EPsssataalqsaggssssvrlsqtvsssQGIRL-------NSDLSALSIRATEYLQRWK 339
P S+ + +S + G+ + + LS L++R E LQ+W+
Sbjct 267 SPIFGDLDIGDSSSLTFASAPQQGANTIQSGVLVVNNNSNTTAGLSVLALRQAECLQKWR 326
Query 340 EIVQFSSKDYSDQMAAQFGIKAPEYMGNHSHYIGGWSSVININEVVNTNLDADSSQASIA 399
EI Q DY QM F + + H Y+GGW+S ++I+EVVNTNL D +QA I
Sbjct 327 EIAQSGKMDYQTQMQKHFNVSPSATLSGHCKYLGGWTSNLDISEVVNTNLTGD-NQADIQ 385
Query 400 GKGISSNSGHTLTYDCGAEHQIIMCVYHAVPMLDWNLTGQAPQLTVTAISDFPQPAFDQL 459
GKG + +G+ + ++ +EH IIMC+YH +P+LDW++ A Q T +D+ P FD +
Sbjct 386 GKGTGTLNGNKVDFE-SSEHGIIMCIYHCLPLLDWSINRIARQNFKTTFTDYAIPEFDSV 444
Query 460 GMQSV-PS---LNLQNNPGRNVSGALGYNLRYWQWKSNIDTVHAGFRAGAAYQSWVAPL- 514
GMQ + PS L++ P S +GY RY K++ID +H F SWV+PL
Sbjct 445 GMQQLYPSEMIFGLEDLPSDPSSINMGYVPRYADLKTSIDEIHGSFIDTLV--SWVSPLT 502
Query 515 DGW---------NVLTSSGAWSYQSMKVRPQQLNSIFVPQVDSANCSVAFDQLLCNVNFQ 565
D + + S +Y KV P +++IF + DS ++ DQLL N F
Sbjct 503 DSYISAYRQACKDAGFSDITMTYNFFKVNPHIVDNIFGVKADS---TINTDQLLINSYFD 559
Query 566 VYAVQNLDRNGLPY 579
+ AV+N D NGLPY
Sbjct 560 IKAVRNFDYNGLPY 573
>gi|496050829|ref|WP_008775336.1| hypothetical protein [Bacteroides sp. 2_2_4]
gi|229448893|gb|EEO54684.1| putative capsid protein (F protein) [Bacteroides sp. 2_2_4]
Length=580
Score = 280 bits (716), Expect = 3e-82, Method: Compositional matrix adjust.
Identities = 203/620 (33%), Positives = 295/620 (48%), Gaps = 81/620 (13%)
Query 1 MSDFNPLNRARISTHRSSFDLSSKKLFTAKVGEILPCYWQIAIPGNKYRISSDWFTRTVP 60
M++ L R T R+ FDLSSK+ FTAK GE+LP +PG+K+ I FTRT P
Sbjct 1 MANIMSLKSLRNKTSRNGFDLSSKRNFTAKPGELLPVKCWEVLPGDKWSIDLKSFTRTQP 60
Query 61 VNTAAYTRIKEYYDFYAVPLRLISRALPQAFTQMTDYItsaasstansstltsVPFVSQT 120
+NTAA+ R++EYYDFY VP L+ TQM D + +P +Q
Sbjct 61 LNTAAFARMREYYDFYFVPYNLLWNKANTVLTQMYD---------NPQHATSYIPSANQA 111
Query 121 LFNAFFQTANAG----------DQPNT----RDDAGLPIVYGSCKLLDMLGYGSMIASNN 166
L G D T ++ G G+ KLL+ LGYG+
Sbjct 112 LAGVMPNVTCKGIADYLNLVAPDVTTTNSYEKNYFGYSRSLGTAKLLEYLGYGNFYTYAT 171
Query 167 PSKAAITKKYLGVDSLNDADNPLVYQTSQTVNALPFLAYQKIYYDFFSNSQWEKHKAYAY 226
TK +PL ++ +N LAYQKIY D +SQWEK +
Sbjct 172 SKNNTWTK------------SPL--SSNLQLNIYGVLAYQKIYADHIRDSQWEKVSPSCF 217
Query 227 NVDYWSGAGNIGLVTD-------------MVQLRYANYPKDYFMGMLPSSQYGSVAVLPs 273
NVDY SG + + D M LRY N+ KD F G+LP QYG A +
Sbjct 218 NVDYLSGTVDSAMTIDSMITGQGFAPFYNMFDLRYCNWQKDLFHGVLPRQQYGDTAAVNV 277
Query 274 issssdsrsLLYYEPsssataalqsaggssssvrlsqtvsssQGIRLNSDLSALSIRATE 333
S+ S + P + + Q + + + L++R E
Sbjct 278 NLSNVLSAQYMVQTPDGDPVGGSPFSSTGVNL----------QTVNGSGTFTVLALRQAE 327
Query 334 YLQRWKEIVQFSSKDYSDQMAAQFGIKAPEYMGNHSHYIGGWSSVININEVVNTNLDADS 393
+LQ+WKEI Q +KDY DQ+ + + E S Y+GG ++ ++INEVVN N+ S
Sbjct 328 FLQKWKEITQSGNKDYKDQIEKHWNVSVGEAYSEMSLYLGGTTASLDINEVVNNNITG-S 386
Query 394 SQASIAGKGISSNSGHTLTYDCGAEHQIIMCVYHAVPMLDWNLTGQAPQLTVTAISDFPQ 453
+ A IAGKG+ +G +++D G + +IMC+YH++P+LD+ P T +DF
Sbjct 387 NAADIAGKGVVVGNGR-ISFDAGERYGLIMCIYHSLPLLDYTTDLVNPAFTKINSTDFAI 445
Query 454 PAFDQLGMQSVPSLNLQN--NPGRNV-SGALGYNLRYWQWKSNIDTVHAGFRAGAAYQSW 510
P FD++GM+SVP ++L N NV S LGY RY +K+++D+ F+ +SW
Sbjct 446 PEFDRVGMESVPLVSLMNPLQSSYNVGSSILGYAPRYISYKTDVDSSVGAFK--TTLKSW 503
Query 511 VAPLDGWNVL----------TSSGAW-SYQSMKVRPQQLNSIFVPQVDSANCSVAFDQLL 559
V D +V+ S G +Y + KV P ++ +F +A+ S+ DQ L
Sbjct 504 VMSYDNQSVINQLNYQDDPNNSPGTLVNYTNFKVNPNCVDPLFAV---AASNSIDTDQFL 560
Query 560 CNVNFQVYAVQNLDRNGLPY 579
C+ F V V+NLD +GLPY
Sbjct 561 CSSFFDVKVVRNLDTDGLPY 580
>gi|490418709|ref|WP_004291032.1| hypothetical protein [Bacteroides eggerthii]
gi|217986636|gb|EEC52970.1| putative capsid protein (F protein) [Bacteroides eggerthii DSM
20697]
Length=578
Score = 273 bits (697), Expect = 1e-79, Method: Compositional matrix adjust.
Identities = 209/617 (34%), Positives = 297/617 (48%), Gaps = 77/617 (12%)
Query 1 MSDFNPLNRARISTHRSSFDLSSKKLFTAKVGEILPCYWQIAIPGNKYRISSDWFTRTVP 60
M++ L R R+ FDLS KK FTAK GE+LP + +PG+ ++I+ FTRT P
Sbjct 1 MANIMSLKSIRNKPSRNGFDLSFKKNFTAKAGELLPVMVKEVLPGDTFKINLKAFTRTQP 60
Query 61 VNTAAYTRIKEYYDFYAVPLRLISRALPQAFTQMTDYItsaass--tansstltsVPFVS 118
VNTAA+ RI+EYYDF+ VP L+ TQM D A S T N +P+++
Sbjct 61 VNTAAFARIREYYDFFFVPYDLLWNKANTVLTQMYDNPQHAVSIDPTRNFVLSGEMPYMT 120
Query 119 Q----TLFNAFFQTANAGDQPNTRDDAGLPIVYGSCKLLDMLGYGSMIASNNPSKAAITK 174
+ NA + D + G S KLL+ LGYG+ +
Sbjct 121 SEAIASYINALSTASALADYKSNY--FGYNRSKSSVKLLEYLGYGNY------------E 166
Query 175 KYLGVDSLNDADNPLVYQTSQTVNALPFLAYQKIYYDFFSNSQWEKHKAYAYNVDYWSGA 234
+L D N A PL+ + + L LAYQKIY DF+ +SQWE+ +NVDY G+
Sbjct 167 SFL-TDDWNTA--PLMANLNHNIFGL--LAYQKIYSDFYRDSQWERVSPSTFNVDYLDGS 221
Query 235 G---NIGLVTDMVQ------LRYANYPKDYFMGMLPSSQYGSVAVLPsissssdsrsLLY 285
+ T+ Q LRY N+ KD F G+LP QYG AV + +L
Sbjct 222 SMNLDNAYSTEFYQNYNFFDLRYCNWQKDLFHGVLPHQQYGETAVASITPDVTGKLTLSN 281
Query 286 YEPsssataalqsaggssssvrlsqtvsssQGIRLNSDLSALSIRATEYLQRWKEIVQFS 345
+ G+S + + DLS L +R E+LQ+WKEI Q
Sbjct 282 FS-----------TVGTSPTTASGTATKNLPAFDTVGDLSILVLRQAEFLQKWKEITQSG 330
Query 346 SKDYSDQMAAQFGIKAPEYMGNHSHYIGGWSSVININEVVNTNLDADSSQASIAGKGISS 405
+KDY DQ+ +G+ + Y+GG SS I+INEV+NTN+ S+ A IAGKG+
Sbjct 331 NKDYKDQLEKHWGVSVGDGFSELCTYLGGVSSSIDINEVINTNITG-SAAADIAGKGVGV 389
Query 406 NSGHTLTYDCGAEHQIIMCVYHAVPMLDWNLTGQAPQLTVTAISDFPQPAFDQLGMQSVP 465
+G + ++ + +IMC+YH +P+LD+ P +D+ P FD++GMQS+P
Sbjct 390 ANGE-INFNSNGRYGLIMCIYHCLPLLDYTTDMLDPAFLKVNSTDYAIPEFDRVGMQSMP 448
Query 466 SLNLQNNPGR---NVSG-ALGYNLRYWQWKSNIDTVHAGFRAGAAYQSWV---------- 511
+ L NP R N SG LGY RY +K+++D GF+ SWV
Sbjct 449 LVQLM-NPLRSFANASGLVLGYVPRYIDYKTSVDQSVGGFK--RTLNSWVISYGNISVLK 505
Query 512 --------APLDGWNVLTSSGAWSYQSMKVRPQQLNSIFVPQV-DSANCSVAFDQLLCNV 562
P++ + S ++ KV P L+ IF Q D N DQ LC+
Sbjct 506 QVTLPNDAPPIEPSEPVPSVAPMNFTFFKVNPDCLDPIFAVQAGDDTNT----DQFLCSS 561
Query 563 NFQVYAVQNLDRNGLPY 579
F + AV+NLD +GLPY
Sbjct 562 FFDIKAVRNLDTDGLPY 578
>gi|494822885|ref|WP_007558293.1| hypothetical protein [Bacteroides plebeius]
gi|198272099|gb|EDY96368.1| putative capsid protein (F protein) [Bacteroides plebeius DSM
17135]
Length=613
Score = 228 bits (580), Expect = 1e-62, Method: Compositional matrix adjust.
Identities = 175/629 (28%), Positives = 299/629 (48%), Gaps = 73/629 (12%)
Query 1 MSDFNPLNRARISTHRSSFDLSSKKLFTAKVGEILPCYWQIAIPGNKYRISSDWFTRTVP 60
M++ + R R+ +DL+ K FTAK G ++P +W +P + + F RT P
Sbjct 8 MANIMSMKSVRNKPTRAGYDLTQKINFTAKAGSLIPVWWTPVLPFDDLNATVKSFVRTQP 67
Query 61 VNTAAYTRIKEYYDFYAVPLRLISRALPQAFTQMTDYItsaasstansstltsVPFVSQT 120
+NTAA+ R++ Y+DFY VP R + P A TQM + A+ + VP +
Sbjct 68 LNTAAFARMRGYFDFYFVPFRQMWNKFPTAITQMRTNLLHASGPVLADN----VPLSDEL 123
Query 121 LFNAFFQTAN-AGDQPNTRDDAGLPIVYGSCKLLDMLGYGSMIASNNPSKAAITKKYLGV 179
+ Q A+ ++++ G + C +L+ LGYG + G
Sbjct 124 PYFTAEQVADYIVSLADSKNQFGYYRAWLVCIILEYLGYGDFYP--------YIVEAAGG 175
Query 180 DSLNDADNPLVYQTSQTVNALPFLAYQKIYYDFFSNSQWEKHKAYAYNVDYWSGAGNIGL 239
+ A P++ + + P AYQKIY DF +QWE+ +N+DY SG+ + L
Sbjct 176 EGATWATRPML--NNLKFSPFPLFAYQKIYADFNRYTQWERSNPSTFNIDYISGSAD-SL 232
Query 240 VTD-----------MVQLRYANYPKDYFMGMLPSSQYGSVAVL--------------Psi 274
D + +RY+N+ +D G +P +QYG + + P+
Sbjct 233 QLDFTVEGFKDSFNLFDMRYSNWQRDLLHGTIPQAQYGEASAVPVSGSMQVVEGPTPPAF 292
Query 275 ssssdsrsLLYYEPsssataalqsaggssssvrlsqtvsssQGIRLNSD----LSALSIR 330
++ D + L + ++ A S R+ + +++ G+ + D +S L++R
Sbjct 293 TTGQDGVAFLNGNVTIQGSSGYLQAQTSVGESRILRFNNTNSGLIVEGDSSFGVSILALR 352
Query 331 ATEYLQRWKEIVQFSSKDYSDQMAAQFGIKAPEYMGNHSHYIGGWSSVININEVVNTNLD 390
E Q+WKE+ S +DY Q+ A +G + + ++G + ++INEVVN N+
Sbjct 353 RAEAAQKWKEVALASEEDYPSQIEAHWGQSVNKAYSDMCQWLGSINIDLSINEVVNNNIT 412
Query 391 ADSSQASIAGKGISSNSGHTLTYDCGAEHQIIMCVYHAVPMLDWNLTGQAPQLTVTAISD 450
+++ A IAGKG S +G ++ ++ G ++ I+MCV+H +P LD+ + T+T + D
Sbjct 413 GENA-ADIAGKGTMSGNG-SINFNVGGQYGIVMCVFHVLPQLDYITSAPHFGTTLTNVLD 470
Query 451 FPQPAFDQLGMQSVPSLNLQNNPGRNVSGA--------LGYNLRYWQWKSNIDTVHAGFR 502
FP P FD++GM+ VP + NP + G GY +Y+ WK+ +D FR
Sbjct 471 FPIPEFDKIGMEQVPVIR-GLNPVKPKDGDFKVSPNLYFGYAPQYYNWKTTLDKSMGEFR 529
Query 503 AGAAYQSWVAPLDGWNVLTSSG----------AWSYQS--MKVRPQQLNSIFVPQVDSAN 550
+ ++W+ P D +L + A S ++ KV P L+++F + AN
Sbjct 530 --RSLKTWIIPFDDEALLAADSVDFPDNPNVEADSVKAGFFKVSPSVLDNLFAVK---AN 584
Query 551 CSVAFDQLLCNVNFQVYAVQNLDRNGLPY 579
+ DQ LC+ F V V++LD NGLPY
Sbjct 585 SDLNTDQFLCSTLFDVNVVRSLDPNGLPY 613
>gi|494308783|ref|WP_007173938.1| hypothetical protein [Prevotella bergensis]
gi|270333035|gb|EFA43821.1| putative capsid protein (F protein) [Prevotella bergensis DSM
17361]
Length=553
Score = 187 bits (474), Expect = 3e-48, Method: Compositional matrix adjust.
Identities = 161/587 (27%), Positives = 262/587 (45%), Gaps = 64/587 (11%)
Query 10 ARISTHRSSFDLSSKKLFTAKVGEILPCYWQIAIPGNKYRISSDWFTRTVPVNTAAYTRI 69
R + +R++FDLS + LFTA G +LP IP + I++ F RT+P+NTAA+ +
Sbjct 11 TRPNRNRNAFDLSQRHLFTAHAGMLLPVLNLDLIPHDHVEINAQDFMRTLPMNTAAFASM 70
Query 70 KEYYDFYAVPLRLISRALPQAFTQMTDYItsaasstansstltsVPFVSQTLFNAFFQTA 129
+ Y+F+ VP + Q T M D+ +SA S ++ VP+ + ++ F +
Sbjct 71 RGVYEFFFVPYHQLWAQFDQFITGMNDFHSSANKSIQGGTSPLQVPYFN---VDSVFNSL 127
Query 130 NAGDQ--PNTRDDAGLPIVYGSCKLLDMLGYGSMIASNNPSKAAITKKYLGVDSLNDADN 187
N G + + DD YG+ +LLD+LGYG S + D+++ N
Sbjct 128 NTGKESGSGSTDDLQYKFKYGAFRLLDLLGYGRKFDSFGTAYP---------DNVSGLKN 178
Query 188 PLVYQTSQTVNALPFLAYQKIYYDFFSNSQWEKHKAYAYNVDYWSGA-GNIGLVTDMVQL 246
L Y S LAY KIY D++ NS +E ++N D + G + +V D+ +L
Sbjct 179 NLDYNCS----VFRILAYNKIYQDYYRNSNYENFDTDSFNFDKFKGGLVDAKVVADLFKL 234
Query 247 RYANYPKDYFMGMLPSSQYGSVAVLPsissssdsrsLLYYEPsssataalqsaggssssv 306
RY N DYF + S L + + + A
Sbjct 235 RYRNAQTDYFTNLRQSQ-------------------LFSFTTAFEDVDNINIAPRDYVKS 275
Query 307 rlsqtvsssQGIRLNS---DLSALSIRATEYLQRWKEIVQFSSKDYSDQMAAQFGIKAPE 363
S + G+ +S D S S+RA + + + + K + DQM A +G++ P+
Sbjct 276 DGSNFTRVNFGVDTDSSEGDFSVSSLRAAFAVDKLLSVTMRAGKTFQDQMRAHYGVEIPD 335
Query 364 YMGNHSHYIGGWSSVININEVVNTNLDADSSQ-------ASIAGKGISSNSGHTLTYDCG 416
+Y+GG+ S + +++V T+ + +AGKG S G + +D
Sbjct 336 SRDGRVNYLGGFDSDMQVSDVTQTSGTTATEYKPEAGYLGRVAGKGTGSGRGR-IVFDA- 393
Query 417 AEHQIIMCVYHAVPMLDWNLTGQAPQLTVTAISDFPQPAFDQLGMQSVPSLNLQN----N 472
EH ++MC+Y VP + ++ T P + D+ P F+ LGMQ + S + + +
Sbjct 394 KEHGVLMCIYSLVPQIQYDCTRLDPMVDKLDRFDYFTPEFENLGMQPLNSSYISSFCTTD 453
Query 473 PGRNVSGALGYNLRYWQWKSNIDTVHAGFRAGAAYQSW-VAPLDGWNVLTSSGAWSYQSM 531
P V LGY RY ++K+ +D H F A SW V+ W T+
Sbjct 454 PKNPV---LGYQPRYSEYKTALDVNHGQFAQSDALSSWSVSRFRRW---TTFPQLEIADF 507
Query 532 KVRPQQLNSIFVPQVDSANCSVAFDQLLCNVNFQVYAVQNLDRNGLP 578
K+ P LNSIF VD N + A D + NF + V ++ +G+P
Sbjct 508 KIDPGCLNSIF--PVD-YNGTEANDCVYGGCNFNIVKVSDMSVDGMP 551
>gi|575094321|emb|CDL65708.1| unnamed protein product [uncultured bacterium]
Length=642
Score = 180 bits (457), Expect = 1e-45, Method: Compositional matrix adjust.
Identities = 181/643 (28%), Positives = 287/643 (45%), Gaps = 100/643 (16%)
Query 16 RSSFDLSSKKLFTAKVGEILPCYWQIAIPGNKYRISSDWFTRTVPVNTAAYTRIKEYYDF 75
R+SFDLS + +FTAKVGE+LPC+ Q PG+ ++SS +FTRT P+ + A+TR++E +
Sbjct 19 RNSFDLSHRNMFTAKVGELLPCFVQELNPGDSVKVSSSYFTRTAPLQSNAFTRLRENVQY 78
Query 76 YAVPLRLISRALPQAFTQMT------DYItsaasstansstltsVPFVSQTLFNAFF--- 126
+ VP + + MT D A+S N T +P V+ +A+
Sbjct 79 FFVPYSALWKYFDSQVLNMTKNANGGDISRIASSLVGNQKVTTQMPCVNYKTLHAYLLKF 138
Query 127 ---QTANAGDQPNTRDDAGLPIVYGSCKLLDMLGYGSMIASNNPSKAAITKKYLGVDSLN 183
T + + G S KLL +LGYG N P + A K + D N
Sbjct 139 INRSTVGSDGSVGPEFNRGCYRHAESAKLLQLLGYG-----NFPEQFANFK--VNNDKHN 191
Query 184 DAD---NPLVYQTSQTVNALPFLAYQKIYYDFFSNSQWEKHKAYAYNVDYWSGAGNIGLV 240
+ + Y S ++ LAY KI D + QW+ + A NVDY + + L
Sbjct 192 QSGQNFKDVTYNNSPYLSIFRLLAYHKICNDHYLYRQWQPYNASLCNVDYLTPNSSSLLS 251
Query 241 TD-----------------MVQLRYANYPKDYFMGMLPSSQYGSVAVLPsissssdsrsL 283
D ++ +R++N P DYF G+LP+SQ+GS +V+ ++ ++
Sbjct 252 IDDALLSIPDDSIKAEKLNLLDMRFSNLPLDYFTGVLPTSQFGSESVVNLNLGNASGSAV 311
Query 284 L------------------YYEPsssataalqsaggssssvrlsqtvsssQGIRLNSDLS 325
L E +++A +S+ +S + S + +N+ LS
Sbjct 312 LNGTTSKDSGRWRTTTGEWEMEQRVASSANGNLKLDNSNGTFISHDHTFSGNVAINTSLS 371
Query 326 A----LSIRATEYLQRWKEIVQFSSKDYSDQMAAQFGIKAPEYMGNHSHYIGGWSSVINI 381
+++R Q++KEI + D+ Q+ A FGIK P+ +S +IGG SS+INI
Sbjct 372 GNLSIIALRNALAAQKYKEIQLANDVDFQSQVEAHFGIK-PDEKNENSLFIGGSSSMINI 430
Query 382 NEVVNTNLDADSSQ---ASIAGKGISSNSGHTLTYDCGAEHQIIMCVYHAVPMLDWNLTG 438
NE +N NL D+ A+ G G +S TY +++ +Y P+LD+ G
Sbjct 431 NEQINQNLSGDNKATYGAAPQGNGSASIKFTAKTYG------VVIGIYRCTPVLDFAHLG 484
Query 439 QAPQLTVTAISDFPQPAFDQLGMQSV---------------PSLNLQNNPGRNVSGALGY 483
L T SDF P D +GMQ + + + ++S GY
Sbjct 485 IDRTLFKTDASDFVIPEMDSIGMQQTFRCEVAAPAPYNDEFKAFRVGDGSSPDMSETYGY 544
Query 484 NLRYWQWKSNIDTVHAGFRAGAAYQSWVAPL-------DGWNVLTSSGAWSYQSMKVRPQ 536
RY ++K++ D + F + +SWV + + WN T +G + RP
Sbjct 545 APRYSEFKTSYDRYNGAF--CHSLKSWVTGINFDAIQNNVWN--TWAGINAPNMFACRPD 600
Query 537 QLNSIFVPQVDSANCSVAFDQLLCNVNFQVYAVQNLDRNGLPY 579
+ ++F+ V S N S DQL + YA +NL R GLPY
Sbjct 601 IVKNLFL--VSSTNNSDD-DQLYVGMVNMCYATRNLSRYGLPY 640
>gi|490477384|ref|WP_004347761.1| capsid protein [Prevotella buccalis]
gi|281300712|gb|EFA93043.1| putative capsid protein (F protein) [Prevotella buccalis ATCC
35310]
Length=552
Score = 174 bits (440), Expect = 1e-43, Method: Compositional matrix adjust.
Identities = 161/601 (27%), Positives = 260/601 (43%), Gaps = 74/601 (12%)
Query 1 MSDFNPLNRA-RISTHRSSFDLSSKKLFTAKVGEILPCYWQIAIPGNKYRISSDWFTRTV 59
MS PL +A R + R++FDLS K LFTA G +LP IP + I + F R +
Sbjct 1 MSKKIPLIKASRANRPRNAFDLSQKHLFTAHAGMLLPVMTLDLIPHDHVSIQATDFMRCL 60
Query 60 PVNTAAYTRIKEYYDFYAVPLRLISRALPQAFTQMTDYItsaasstansstltsVP-FVS 118
P+N+AA+ ++ Y+F+ VP + Q T M DY + S S + +P F
Sbjct 61 PMNSAAFMSMRSVYEFFFVPYSQLWHPFDQFITGMNDYRSVLQSDLYKSKSPLVIPSFKR 120
Query 119 QTLFNAFFQTANAG---DQPNTRDDAGLPIVYGSCKLLDMLGYGSMIASNNPSKAAITKK 175
+ L+ F A G Q N D G + +LLD+LGYG + ++ S+ K
Sbjct 121 KELYELF--NAPGGFLNQQSNQPDIFGFKSRFNFLRLLDLLGYGVYVNADGSSRIDAFSK 178
Query 176 YLGVDSLNDADNPLVYQTSQTVNALPFLAYQKIYYDFFSNSQWEKHKAYAYNVDYWSGA- 234
L ++ ++ AYQKIY DF+ N+ +E ++++D + +
Sbjct 179 LL--------------DDTEKLSIFRLAAYQKIYSDFYRNTTYEAVDVSSFSLDNITDSI 224
Query 235 GNIGLVTDMVQLRYANYPKDYFMGMLPSSQYGSVAVLPsissssdsrsLLYYEPsssata 294
I LRY N DYF + P+ P + S + Y P ++ +
Sbjct 225 SAINAFKRFGTLRYRNAQLDYFTNLRPT---------PLFDLDNPSLNSFYNTPGNADSV 275
Query 295 alqsaggssssvrlsqtvsssQGIRLNSDL-SALSIRATEYLQRWKEIVQFSSKDYSDQM 353
++ S + +L+SDL + SIR L + I Q + K Y++Q+
Sbjct 276 SIDSDSNAV-------------NFQLDSDLLTVQSIRNAFALDKLMRITQRAGKTYAEQI 322
Query 354 AAQFGIKAPEYMGNHSHYIGGWSSVININEVVNTNLDADSSQ-----------ASIAGKG 402
A FG + E +YIGG+ S I + +V + S + + GK
Sbjct 323 KAHFGFEVSEGRDGRVNYIGGFDSNIQVGDVTQMSGTTASPEQGVSIKHGGYLGRVTGKA 382
Query 403 ISSNSGHTLTYDCGAEHQIIMCVYHAVPMLDWNLTGQAPQLTVTAISDFPQPAFDQLGMQ 462
S SGH + +D EH I+MC+Y VP + ++ T P +T + DF P F+ LGMQ
Sbjct 383 QGSGSGH-IEFDA-HEHGILMCIYSLVPDMQYDATRIDPFVTKLSRGDFFMPEFEDLGMQ 440
Query 463 SVPSLNLQNNPGRNVSGALGYNLRYWQWKSNIDTVHAGFRAGAAYQSWVAPLDGWNVLTS 522
+ + + ++ G+ RY ++K+++D H F G PL W V
Sbjct 441 PLQTRYI-SDIRTQTEKFKGWQPRYSEYKTSLDINHGQFANG-------QPLSYWTVGRG 492
Query 523 SGAWSYQ-----SMKVRPQQLNSIFVPQVDSANCSVAFDQLLCNVNFQVYAVQNLDRNGL 577
+ + S+K+ P+ L+SIF + + D + F V V ++ NG
Sbjct 493 RAGETLETFDIASLKINPKWLDSIFAVNYNGTQIT---DCVFGGCQFNVQKVSDMSENGE 549
Query 578 P 578
P
Sbjct 550 P 550
>gi|647452987|ref|WP_025792807.1| hypothetical protein [Prevotella histicola]
Length=584
Score = 172 bits (437), Expect = 3e-43, Method: Compositional matrix adjust.
Identities = 174/625 (28%), Positives = 265/625 (42%), Gaps = 98/625 (16%)
Query 6 PLNRARISTHRSSFDLSSKKLFTAKVGEILPC-YWQIAIPGNKYRISSDWFTRTVPVNTA 64
P + R++ R+ FDLSS+++F+AK G++LP W++ P ++ S RT +NTA
Sbjct 2 PAPKPRLA--RNGFDLSSRRIFSAKAGQLLPIGCWEVN-PSEHFKFSVQDLVRTTTLNTA 58
Query 65 AYTRIKEYYDFYAVPLRLISRALPQAFTQMTDYItsaasstansstltsVPFVSQTLFNA 124
+Y R+KEYY F+ V R +L Q F Q + S L V T +N
Sbjct 59 SYARMKEYYHFFFVSYR----SLWQWFDQFI------VGTNNPHSALNGVKKNGTTNYNQ 108
Query 125 FFQTANAGD--------QPNTRDDAGLPIVYGSCKLLDMLGYGSMIASN--NPSKAAITK 174
+ D + + D G G+ KLL+ML YG N +
Sbjct 109 ICSSVPTFDLGKLITRLKTSDMDSQGFNYSEGAAKLLNMLNYGVTNKGKFMNLENLITST 168
Query 175 KYLGVDSLNDADNPLVYQTSQTVNALPFLAYQKIYYDFFSNSQWEKHKAYAYNVDYWSGA 234
YL S +D + +Y V+ LAYQKI+ DF+ N W ++NVD ++
Sbjct 169 SYL--PSKDDKEPSSIYACK--VSPFRLLAYQKIFNDFYRNQDWTPSDVRSFNVDDYADD 224
Query 235 GNIGLVTDM----VQLRYANYPKDYFMGMLPSSQYG-SVAVLPsissssdsrsLLYYEPs 289
N+ + D+ Q+RY Y KD+ M P+ Y + LP + + L
Sbjct 225 SNLTIEPDVALKFCQMRYRPYAKDWLTSMKPTPNYSDGIFNLPEYVRGNGNVIL------ 278
Query 290 ssataalqsaggssssvrlsqtvsssQGIRLNSDLSALSIRATEYLQRWKEIVQFSSK-D 348
+ S +VS G S S +RA L + E + ++ D
Sbjct 279 ---------------TNNKSGSVSLDSGTVSPSSFSVNDLRAAFALDKMLEATRRANGLD 323
Query 349 YSDQMAAQFGIKAPEYMGNHSHYIGGWSSVININEVVNTNLDA--DSSQASI---AGKGI 403
Y+ Q+ A FG K PE N + ++GG+ + I ++EVV+TN +A D S ASI GKGI
Sbjct 324 YASQIEAHFGFKVPESRANDARFLGGFDNSIVVSEVVSTNGNAASDGSHASIGDLGGKGI 383
Query 404 SSNSGHTLTYDCGAEHQIIMCVYHAVPMLDWNLTGQAPQLTVTAISDFPQPAFDQLGMQS 463
S S T+ +D EH IIMC+Y P ++N + P F QP F LG Q+
Sbjct 384 GSMSSGTIEFDS-TEHGIIMCIYSVAPQSEYNASYLDPFNRKLTREQFYQPEFADLGYQA 442
Query 464 VPSLNL-QNNPGRNVSGA-----------LGYNLRYWQWKSNIDTVHAGFRAGAAYQSWV 511
+ +L + G N A LGY +RY ++K+ D V F +G + W
Sbjct 443 LIGSDLICSTLGMNEKQAGFSDIELNNNLLGYQVRYNEYKTARDLVFGDFESGKSLSYWC 502
Query 512 APL-------------------DGWNVLTSSGAWSYQSMKVRPQQLNSIFVPQVDSANCS 552
P + + WS ++ + P +N IF+ +
Sbjct 503 TPRFDFGYGDTEKKIAPENKGGADYRKKGNRSHWSSRNFYINPNLVNPIFL------TSA 556
Query 553 VAFDQLLCNVNFQVYAVQNLDRNGL 577
V D + N V AV+ + GL
Sbjct 557 VQADHFIVNSFLDVKAVRPMSVTGL 581
>gi|517172762|ref|WP_018361580.1| hypothetical protein [Prevotella nanceiensis]
Length=568
Score = 166 bits (419), Expect = 6e-41, Method: Compositional matrix adjust.
Identities = 161/604 (27%), Positives = 264/604 (44%), Gaps = 75/604 (12%)
Query 6 PLNRARISTH-RSSFDLSSKKLFTAKVGEILPCYWQIAIPGNKYRISSDWFTRTVPVNTA 64
PL + +T R++FD+S + LFTA G +LP +P + I++ F RT+P+N+A
Sbjct 7 PLIKPSKATRPRNAFDISQRHLFTAPAGALLPVLSLDLLPHDHVEINASDFMRTLPMNSA 66
Query 65 AYTRIKEYYDFYAVPLRLISRALPQAFTQMTDYItsaasstansstltsVPFVSQTLFNA 124
A+ ++ Y+FY VP + + Q T M+DY +S + + + V F Q L +
Sbjct 67 AFMSMRGVYEFYFVPYKQLWSGFDQFITGMSDYKSSFMYAFKGKTPPSCVSFDVQKLVD- 125
Query 125 FFQTANAGDQPNTRDDAGLPIVYGSCKLLDMLGYGSMIASNNPSKAAITKKYLGVDSLND 184
+ +T A +D G G ++LD+LGYG KY N
Sbjct 126 WCKTNTA------KDIHGFDKNKGVYRILDLLGYG---------------KY-----ANS 159
Query 185 ADNPLVYQTSQTV-NALPF--LAYQKIYYDFFSNSQWEKHKAYAYNVDYWSGAGNIGLVT 241
A P TS T+ PF LAYQKIY DF+ N+ +E+++ ++NVD + G+G +
Sbjct 160 AGVPYTNPTSTTMGKCTPFRGLAYQKIYNDFYRNTTYEEYQLESFNVDMFYGSGKVKETI 219
Query 242 -------DMVQLRYANYPKDYFMGMLPSSQYGSVAVLPsissssdsrsLLYYEPsssata 294
D LRY N KD + P+ + S+ + S ++ P+ +
Sbjct 220 PNEPWDYDWFTLRYRNAQKDLLTNVRPTPLF-SIDDFNPQFFTGGSDIVMEKGPNVTGGT 278
Query 295 alqsaggssssvrlsqtvsssQGIRLNSDLSALSIRATEYLQRWKEIVQFSSKDYSDQMA 354
L + S+ + +S IR L++ + + K Y +QM
Sbjct 279 HEYRDSVVIVGKNLKENGVDSK----RTMISVADIRNAFALEKLASVTMRAGKTYKEQME 334
Query 355 AQFGIKAPEYMGNHSHYIGGWSSVININEVVNTNLDA-----DSS----QASIAGKGISS 405
A FGI E YIGG+ S I + +V ++ D+S GK S
Sbjct 335 AHFGISVEEGRDGRCTYIGGFDSNIQVGDVTQSSGTTVTGTKDTSFGGYLGRTTGKATGS 394
Query 406 NSGHTLTYDCGAEHQIIMCVYHAVPMLDWNLTGQAPQLTVTAISDFPQPAFDQLGMQSVP 465
SGH + +D EH I+MC+Y VP + ++ P + DF P F+ LGMQ +
Sbjct 395 GSGH-IRFDA-KEHGILMCIYSLVPDVQYDSKRVDPFVQKIERGDFFVPEFENLGMQPLF 452
Query 466 SLNLQNNPGRNVS-------GALGYNLRYWQWKSNIDTVHAGFRAGAAYQSWVAPLDGWN 518
+ N+ N + GA G+ RY ++K+ +D H F +Q PL W
Sbjct 453 AKNISYKYNNNTANSRIKNLGAFGWQPRYSEYKTALDINHGQF----VHQE---PLSYWT 505
Query 519 VLTSSGA----WSYQSMKVRPQQLNSIFVPQVDSANCSVAFDQLLCNVNFQVYAVQNLDR 574
V + G ++ + K+ P+ L+ +F + + DQ+ F + V ++
Sbjct 506 VARARGESMSNFNISTFKINPKWLDDVFAVNYNGTELT---DQVFGGCYFNIVKVSDMSI 562
Query 575 NGLP 578
+G+P
Sbjct 563 DGMP 566
>gi|494306153|ref|WP_007173049.1| hypothetical protein [Prevotella bergensis]
gi|270333881|gb|EFA44667.1| putative capsid protein (F protein) [Prevotella bergensis DSM
17361]
Length=519
Score = 164 bits (416), Expect = 8e-41, Method: Compositional matrix adjust.
Identities = 147/561 (26%), Positives = 245/561 (44%), Gaps = 61/561 (11%)
Query 34 ILPCYWQIAIPGNKYRISSDWFTRTVPVNTAAYTRIKEYYDFYAVPLRLISRALPQAFTQ 93
+LP IP + I++ F RT+P+NTAA+ ++ Y+F+ VP + Q T
Sbjct 2 LLPVLNLDLIPHDHVEINAQDFMRTLPMNTAAFASMRGVYEFFFVPYHQLWAQFDQFITG 61
Query 94 MTDYItsaasstansstltsVPFVSQTLFNAFFQTANAGDQPNTRDDAGLPIVYGSCKLL 153
M D+ +SA S ++ VP+ + L + F P+ +DD YG+ +LL
Sbjct 62 MNDFHSSANKSIQGGTSPLQVPYFN--LESVFKNIIERDSTPSFQDDLQYRFKYGAFRLL 119
Query 154 DMLGYGSMIASNNPSKAAITKKYLGVDSLNDADNPLVYQTSQTVNALPFLAYQKIYYDFF 213
D+LGYG S + D+++ N L Y S LAY KIY D++
Sbjct 120 DLLGYGRKFDSFGTAYP---------DNVSGLKNNLDYNCS----VFRVLAYNKIYQDYY 166
Query 214 SNSQWEKHKAYAYNVDYWSGA-GNIGLVTDMVQLRYANYPKDYFMGMLPSSQYGSVAVLP 272
NS +E ++N D + G + +V D+ +LRY N DYF + S
Sbjct 167 RNSNYENFDTDSFNFDKFKGGLVDAKVVADLFKLRYRNAQTDYFTNLRQSQ--------- 217
Query 273 sissssdsrsLLYYEPsssataalqsaggssssvrlsqtvsssQGIRLNSDL---SALSI 329
L + P S L + S + + ++++L S S+
Sbjct 218 ----------LFTFIPEFSDDEHLNFDRDQYADQSKSNFTQLNFPVDVDNNLGYFSVSSL 267
Query 330 RATEYLQRWKEIVQFSSKDYSDQMAAQFGIKAPEYMGNHSHYIGGWSSVININEVVNTNL 389
R+ + + + + K + DQM A +G++ P+ +Y+GG+ S + +++V T+
Sbjct 268 RSAFAVDKLLSVTMRAGKTFQDQMRAHYGVEIPDSRDGRVNYLGGFDSDLQVSDVTQTSG 327
Query 390 DADSSQ-------ASIAGKGISSNSGHTLTYDCGAEHQIIMCVYHAVPMLDWNLTGQAPQ 442
+ IAGKG S G + +D EH ++MC+Y VP + ++ T P
Sbjct 328 TTATEYKPEAGYLGRIAGKGTGSGRGR-IVFD-AKEHGVLMCIYSLVPQIQYDCTRLDPM 385
Query 443 LTVTAISDFPQPAFDQLGMQSVPSLNLQN----NPGRNVSGALGYNLRYWQWKSNIDTVH 498
+ DF P F+ LGMQ + S + + +P V LGY RY ++K+ +D H
Sbjct 386 VDKLDRFDFFTPEFENLGMQPLNSSYISSFCTPDPKNPV---LGYQPRYSEYKTALDINH 442
Query 499 AGFRAGAAYQSW-VAPLDGWNVLTSSGAWSYQSMKVRPQQLNSIFVPQVDSANCSVAFDQ 557
F A SW V+ W T+ K+ P LNS+F + N + + D
Sbjct 443 GQFAQNDALSSWSVSRFRRW---TTFPQLEIADFKIDPGCLNSVFPVEF---NGTESTDC 496
Query 558 LLCNVNFQVYAVQNLDRNGLP 578
+ NF + V ++ +G+P
Sbjct 497 VFGGCNFNIVKVSDMSVDGMP 517
Lambda K H a alpha
0.319 0.133 0.409 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 4256619118725