bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: All non-redundant GenBank CDS translations+PDB+SwissProt+PIR+PRF
excluding environmental samples from WGS projects
49,011,213 sequences; 17,563,301,199 total letters
Query= Contig-19_CDS_annotation_glimmer3.pl_2_5
Length=582
Score E
Sequences producing significant alignments: (Bits) Value
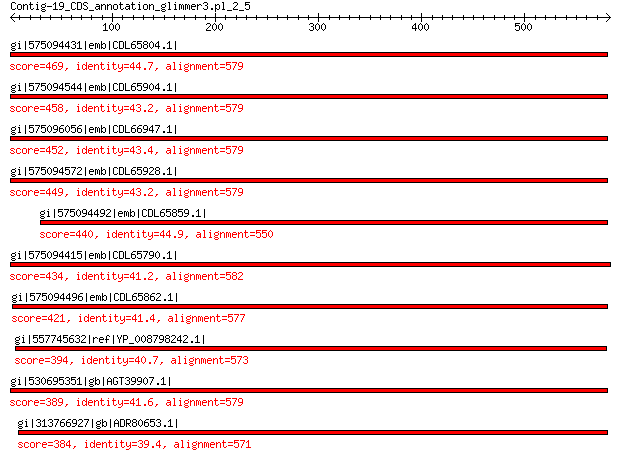
gi|575094431|emb|CDL65804.1| unnamed protein product 469 2e-155
gi|575094544|emb|CDL65904.1| unnamed protein product 458 3e-151
gi|575096056|emb|CDL66947.1| unnamed protein product 452 1e-148
gi|575094572|emb|CDL65928.1| unnamed protein product 449 1e-147
gi|575094492|emb|CDL65859.1| unnamed protein product 440 3e-144
gi|575094415|emb|CDL65790.1| unnamed protein product 434 1e-141
gi|575094496|emb|CDL65862.1| unnamed protein product 421 2e-136
gi|557745632|ref|YP_008798242.1| major capsid protein 394 2e-126
gi|530695351|gb|AGT39907.1| major capsid protein 389 2e-124
gi|313766927|gb|ADR80653.1| putative major coat protein 384 1e-122
>gi|575094431|emb|CDL65804.1| unnamed protein product [uncultured bacterium]
Length=560
Score = 469 bits (1208), Expect = 2e-155, Method: Compositional matrix adjust.
Identities = 259/591 (44%), Positives = 350/591 (59%), Gaps = 47/591 (8%)
Query 1 MNRNNERHFNQVPETHVSRTRFKRDQNILTTFDSGKLIPFYVDEVLPGDTFSVDTAAIIR 60
MNRN+ +F + P +SR+RF R + L TFD+G+++P YVDEVLPGDTF +D AIIR
Sbjct 1 MNRNSNFNFARNPGVSLSRSRFNRTSDRLDTFDTGEIVPIYVDEVLPGDTFELDMTAIIR 60
Query 61 MTTPKYPVMDDAYIDFYYFYCPNRILWDNFKRFMGEADDAPWMPTKTYKVPKIIIDNEEG 120
+TP +PVMD++++D Y+F+ PNR+ W++++ MGE W Y VP++ G
Sbjct 61 GSTPIFPVMDNSFLDVYFFFVPNRLTWEHWRELMGENRTTAWTQPVDYSVPQVTA--PAG 118
Query 121 GAVRAYPDESTILDYMGVPPKAIPVGGTGRIEINALPVRAYVKIWNEFFRDQNVGNPAVL 180
G +E ++ D+MG+P K I +NALP RAY I+NEFFR+QN+ NP +
Sbjct 119 GW-----EELSLADHMGIPTKV------DNISVNALPFRAYGLIYNEFFRNQNLTNPTQV 167
Query 181 NTGDEDEAYRAGSGNESEVSEEKILEYAHIGGYCLPVNRFHDYFSSCLPYPQRGPEVTIA 240
D + A + N ++V ++A G CL +F DYF+ LP PQ+G V I
Sbjct 168 EVTDANIAGK----NPNDVKNSN--DWAITGAKCLKSAKFFDYFTGALPQPQKGEPVEIN 221
Query 241 LTGN----------APLRAYSEKDLNNRKIGTGFFNNE--YNTGIVNHTNISFTKEGTKF 288
L + PL S D + + N + Y G+V +EG
Sbjct 222 LASSWLPVGIGDYHGPLDKVSNSDTLTWESPSSEGNTKRTYALGMVQ-------QEG--- 271
Query 289 SVNKNNNGNTAPLVNGQYIQTMSQDDANFFDAWLGTDLSNIEAATINQLRQAFAVQHYYE 348
VN N N G + ++ + A + W + AAT+NQLRQAF VQ E
Sbjct 272 EVNPNGLKNFETKAGGSFSESGAV-AAYPTNLWASPVTA---AATVNQLRQAFQVQKLLE 327
Query 349 ALARGGSRYREQVRALFGVSISDKTVQIPEYLGGGRYHVNMNQIVQTSGQESNYGTPIGE 408
ARGG+RYRE ++ FGV+ SD +QIPEYLGG + +N++Q+VQTS S +P G
Sbjct 328 KDARGGTRYREILKNHFGVTTSDARMQIPEYLGGCKVPINVSQVVQTSA--STDASPQGN 385
Query 409 TGAMSVTPINESSFTKSFEEHGFVIGVMCVRHDHSYQQGLERFWSRSDRLDYYFPQFANL 468
T A+SVTP ++S FTKSF+EHGF+IGV R SYQQG+ER WSR DRLDYYFP AN+
Sbjct 386 TAAISVTPFSKSMFTKSFDEHGFIIGVATARTAQSYQQGIERMWSRKDRLDYYFPVLANI 445
Query 469 GEQPVKKKEIMLTGKSTDEETFGYQEAWADYRMKPNRVSGKMRSNAEGTLDFWHYADNYA 528
GEQ + KEI G + D+E FGYQEAWADYR KPN + G+ RSNA+ +LD WHY +Y
Sbjct 446 GEQAILNKEIYAQGNAKDDEAFGYQEAWADYRYKPNTICGRFRSNAQQSLDAWHYGQDYD 505
Query 529 TVPTLSQEWMKEGKNEIARTLIVENEPQFFGAIRVMNKTTRCMPLYSVPGL 579
+PTLS +WM++ E+ RTL V+ EP F R KT R MPLYS+PGL
Sbjct 506 KLPTLSTDWMEQSDIEMKRTLAVQTEPDFIANFRFNCKTVRVMPLYSIPGL 556
>gi|575094544|emb|CDL65904.1| unnamed protein product [uncultured bacterium]
Length=551
Score = 458 bits (1179), Expect = 3e-151, Method: Compositional matrix adjust.
Identities = 250/593 (42%), Positives = 361/593 (61%), Gaps = 60/593 (10%)
Query 1 MNRNNERHFNQVPETHVSRTRFKRDQNILTTFDSGKLIPFYVDEVLPGDTFSVDTAAIIR 60
MNRN E HF+++P +SR++F R ++ TTF+ G LIPFY+DEVLPGDTF+V ++ +IR
Sbjct 1 MNRNVESHFSRLPSVDISRSQFDRSSSLKTTFNVGDLIPFYIDEVLPGDTFNVKSSKVIR 60
Query 61 MTTPKYPVMDDAYIDFYYFYCPNRILWDNFKRFMGEADDAPWMPTKTYKVPKIIIDNEEG 120
M + P+MD+ Y+D YYF+ PNR++W ++++F GE ++ W+PT Y+VP++ G
Sbjct 61 MQSLVTPIMDNIYLDTYYFFVPNRLVWSHWQQFNGENTESAWLPTTEYQVPQVTAP-ANG 119
Query 121 GAVRAYPDESTILDYMGVPPKAIPVGGTG-RIEINALPVRAYVKIWNEFFRDQNVGNPAV 179
++ TI DY G+P TG +NALP RAY I NE+FRD+N+ +P
Sbjct 120 WSI------GTIADYFGIP--------TGVACSVNALPFRAYALICNEWFRDENLSDPLN 165
Query 180 LNTGDEDEAYRAGSGNESEVSEEKILEYAHIGGYCLPVNRFHDYFSSCLPYPQRGPEVTI 239
+ D A GS ++ +++ I++ GG ++HDYF+SCLP PQ+GP+V +
Sbjct 166 IPISD---ATVVGSNGDNYITD--IVK----GGMPFKACKYHDYFTSCLPAPQKGPDVLL 216
Query 240 ALTGNAPLRAYSEKDLNNRKIGTGFFNNEYNTGIVNHTNISFTK-----------EGTKF 288
L+ + S+ ++ + ++Y V+ N+S T EG +
Sbjct 217 PLSSSPVPVTTSDTMVDPLQY------SKYPMAGVDSWNLSPTLMRNIIRPFEGVEGANY 270
Query 289 SVNKNNNGNTAPLVNGQYIQTMSQDDANFFDAWLGTDLSNIEAATINQLRQAFAVQHYYE 348
V+ Q+ + DA F L +L N AA+INQLR AF +Q YE
Sbjct 271 QVH-------------QFTGDIPTIDA-FRPLNLVANLQNATAASINQLRLAFQIQRLYE 316
Query 349 ALARGGSRYREQVRALFGVSISDKTVQIPEYLGGGRYHVNMNQIVQTSGQESNYGTPIGE 408
ARGG+RY E +++ FGV+ D +Q PEYLGG R +N+NQ++Q S E+ +P G
Sbjct 317 RDARGGTRYIEILKSHFGVTSPDARLQRPEYLGGNRIPININQVLQQS--ETTSTSPQGN 374
Query 409 TGAMSVTPINESSFTKSFEEHGFVIGVMCVRHDHSYQQGLERFWSRSDRLDYYFPQFANL 468
S+T + F KSF EHGFVIG+M R+DH+YQQGLERFWSR DR DYY+P FA++
Sbjct 375 PVGQSLTTDTNADFVKSFVEHGFVIGLMVARYDHTYQQGLERFWSRKDRFDYYWPVFAHI 434
Query 469 GEQPVKKKEIMLTGKSTDEETFGYQEAWADYRMKPNRVSGKMRSNAEGTLDFWHYADNYA 528
GEQ V KEI +G + D+E FGYQEA+ADYR KP+RV+G+MRS A +LD WH AD+YA
Sbjct 435 GEQAVLNKEIYTSGTAVDDEVFGYQEAYADYRYKPSRVTGEMRSAAPQSLDVWHLADDYA 494
Query 529 TVPTLSQEWMKEGKNEIARTLIVEN--EPQFFGAIRVMNKTTRCMPLYSVPGL 579
++P+LS W++E + + R L V + Q F I + N++TR MP+YSVPGL
Sbjct 495 SLPSLSDSWIRESASTVDRVLAVSSNVSAQLFCDIYIQNRSTRPMPMYSVPGL 547
>gi|575096056|emb|CDL66947.1| unnamed protein product [uncultured bacterium]
Length=570
Score = 452 bits (1164), Expect = 1e-148, Method: Compositional matrix adjust.
Identities = 251/594 (42%), Positives = 352/594 (59%), Gaps = 44/594 (7%)
Query 1 MNRNNERHFNQVPETHVSRTRFKRDQNILTTFDSGKLIPFYVDEVLPGDTFSVDTAAIIR 60
MNRN E HF+ +P +SR+RF R +I TTF++G ++PF+++EVLPGDTFSVD++ ++R
Sbjct 2 MNRNTESHFSLLPHVDISRSRFDRSSSIKTTFNAGDVVPFFLEEVLPGDTFSVDSSKVVR 61
Query 61 MTTPKYPVMDDAYIDFYYFYCPNRILWDNFKRFMGEADDAPWMPTKTYKVPKIIIDNEEG 120
M T P+MD+ Y+D YYF+ PNR++W ++K F GE +++ W+P Y +P++ + G
Sbjct 62 MQTLLTPMMDNVYLDTYYFFVPNRLVWQHWKEFCGENNESAWIPQTEYAIPQL--KSPVG 119
Query 121 GAVRAYPDESTILDYMGVPPKAIPVGGTGRIEINALPVRAYVKIWNEFFRDQNVGNPAVL 180
G + TI DY G+P G + ++ALP RAY I NE+FRD+N+ +P V+
Sbjct 120 GF-----EVGTIADYFGLPT------GVANLSVSALPFRAYALIMNEWFRDENLMDPLVV 168
Query 181 NTGDEDEAYRAGSGNESEVSEEKILEYAHIGGYCLPVNRFHDYFSSCLPYPQRGPEVTIA 240
T D+A G V++ GG ++HDYF+S LP PQ+GP+V I
Sbjct 169 PT---DDATVTGVNTGIFVTD------VAKGGKPFVAAKYHDYFTSALPAPQKGPDVVI- 218
Query 241 LTGNAPLRAYSEKDLNNRKIGTGFFNNEYNTGIVNHTNISFTKEGTKFSVNKNNNGNTAP 300
P+ + ++ G + + I N + S +GT+ +
Sbjct 219 -----PVASAGNYNVVGNGKGLALSDGSKMSIICNGLSGS-NGQGTELFASGILGSQVGS 272
Query 301 LVNGQYIQTMSQDDANF---FDAWLGTDLSN----------IEAATINQLRQAFAVQHYY 347
++ D A LG +L N AATINQLR AF +Q +Y
Sbjct 273 SGGFGSGSSLRGDGIILGVPTAAQLGNNLENSGLIAIASGNAAAATINQLRMAFQIQKFY 332
Query 348 EALARGGSRYREQVRALFGVSISDKTVQIPEYLGGGRYHVNMNQIVQTSGQESNYGTPIG 407
E ARGGSRY E +R+ FGV+ D +Q EYLGG R +N+NQ++Q SG S TP G
Sbjct 333 EKQARGGSRYTEVIRSFFGVTSPDARLQRSEYLGGNRIPININQVIQQSGTGSASTTPQG 392
Query 408 ETGAMSVTPINESSFTKSFEEHGFVIGVMCVRHDHSYQQGLERFWSRSDRLDYYFPQFAN 467
MS T S FTKSF EHGF+IGVMC R+DH+YQQG++R WSR D+ DYY+P F+N
Sbjct 393 TVVGMSQTTDTHSDFTKSFTEHGFIIGVMCARYDHTYQQGIDRMWSRKDKFDYYWPVFSN 452
Query 468 LGEQPVKKKEIMLTGKSTDEETFGYQEAWADYRMKPNRVSGKMRSNAEGTLDFWHYADNY 527
+GEQ +K KEI G +TD+E FGYQEAWA+YR KP+RV+G+MRS+ +LD WH AD+Y
Sbjct 453 IGEQAIKNKEIYAQGNATDDEVFGYQEAWAEYRYKPSRVTGEMRSSYAQSLDVWHLADDY 512
Query 528 ATVPTLSQEWMKEGKNEIARTLIV--ENEPQFFGAIRVMNKTTRCMPLYSVPGL 579
+ +P+LS EW++E + R L V +N QFF I V N TR MP+YS+PGL
Sbjct 513 SKLPSLSDEWIREDAKTLNRVLAVSDQNSNQFFADIYVKNLCTRPMPMYSIPGL 566
>gi|575094572|emb|CDL65928.1| unnamed protein product [uncultured bacterium]
Length=556
Score = 449 bits (1156), Expect = 1e-147, Method: Compositional matrix adjust.
Identities = 250/595 (42%), Positives = 348/595 (58%), Gaps = 59/595 (10%)
Query 1 MNRNNERHFNQVP-ETHVSRTRFKRDQNILTTFDSGKLIPFYVDEVLPGDTFSVDTAAII 59
MNRN E HF + P +SR+ F R ++ TF++G++IPF+++EVLPGDTF V T+ +I
Sbjct 1 MNRNVESHFAKNPTNIDISRSTFDRSSSVKLTFNTGEIIPFFIEEVLPGDTFKVKTSKVI 60
Query 60 RMTTPKYPVMDDAYIDFYYFYCPNRILWDNFKRFMGEADDAPWMPTKTYKVPKIIIDNEE 119
R+ T P+MD+ Y+D YYF+ PNR++W+++K F GE + W+P Y++P++ E
Sbjct 61 RLQTLLTPMMDNIYLDTYYFFVPNRLVWEHWKEFNGENTQSAWIPEVEYQIPQLTA--PE 118
Query 120 GGAVRAYPDESTILDYMGVPPKAIPVGGTGRIEINALPVRAYVKIWNEFFRDQNVGNPAV 179
GG + T+ DY G+P G I +NALP RAY + NE+FRDQN+ +P
Sbjct 119 GGW-----NIGTLADYFGIPT------GVSGISVNALPFRAYALVCNEWFRDQNLSDPLN 167
Query 180 LNTGDEDEAYRAGSGNESEVSEEKILEYAHIGGYCLPVNRFHDYFSSCLPYPQRGPEVTI 239
+ GD + V+ + GG ++HDYF+SCLP PQ+GP+VTI
Sbjct 168 IPVGD---------ATVTGVNTGTFITDVVKGGLPYTAAKYHDYFTSCLPAPQKGPDVTI 218
Query 240 ALTG--NAPLRAYSEKDLNN--RKIGTGFFNNE----YNTGIVNHTNISFTKEGTKFSVN 291
+T N P+ +E + G G N+E Y G + S + + V
Sbjct 219 PVTSGHNLPVMFLNETHDAGPYKPFGVGIQNSELRNFYGFGSGSSGATSTSDTSSTVEVG 278
Query 292 KNNNGNTAPLVNGQYIQTMSQDDANFFDAWLGTDLSNIEA-----ATINQLRQAFAVQHY 346
+ G GQ NF W T++ +E+ ATINQLR AF +Q
Sbjct 279 SDGTGI------GQ----------NF---WTPTNMWAVESGDVGMATINQLRLAFQLQKL 319
Query 347 YEALARGGSRYREQVRALFGVSISDKTVQIPEYLGGGRYHVNMNQIVQTSGQESNYGTPI 406
YE ARGG+RY E +R+ FGV D +Q PEYLGG R +N+NQI+Q S +S +P+
Sbjct 320 YEKDARGGTRYTEIIRSHFGVVSPDSRLQRPEYLGGNRIPINVNQIIQQS--QSTEQSPL 377
Query 407 GETGAMSVTPINESSFTKSFEEHGFVIGVMCVRHDHSYQQGLERFWSRSDRLDYYFPQFA 466
G MSVT S F KSF EHG++IG++ R+DH+YQQGL+R WSR DR D+Y+P A
Sbjct 378 GALAGMSVTTDKNSDFIKSFVEHGYIIGLVVARYDHTYQQGLDRMWSRKDRFDFYWPVLA 437
Query 467 NLGEQPVKKKEIMLTGKSTDEETFGYQEAWADYRMKPNRVSGKMRSNAEGTLDFWHYADN 526
N+GEQ V KEI + G TD+E FGYQEAWA+YR KPNRV G+MRS+A +LD WH D+
Sbjct 438 NIGEQAVLNKEIYIDGSDTDDEVFGYQEAWAEYRYKPNRVCGEMRSSAPQSLDVWHLGDD 497
Query 527 YATVPTLSQEWMKEGKNEIARTLIVEN--EPQFFGAIRVMNKTTRCMPLYSVPGL 579
Y+++P LS W++E K + R L V + Q F I + NK TR MP+YS+PGL
Sbjct 498 YSSLPYLSDSWIREDKTNVDRVLAVTSSVSDQLFADIYICNKATRPMPMYSIPGL 552
>gi|575094492|emb|CDL65859.1| unnamed protein product [uncultured bacterium]
Length=551
Score = 440 bits (1131), Expect = 3e-144, Method: Compositional matrix adjust.
Identities = 247/557 (44%), Positives = 328/557 (59%), Gaps = 47/557 (8%)
Query 30 TTFDSGKLIPFYVDEVLPGDTFSVDTAAIIRMTTPKYPVMDDAYIDFYYFYCPNRILWDN 89
TTF+ G LIPFYVDE+LPGDTFS+DT+ ++RM + PVMD+ Y+D Y+F+ PNR+ W +
Sbjct 31 TTFNVGDLIPFYVDEILPGDTFSIDTSKVVRMQSLLTPVMDNIYLDTYFFFVPNRLTWSH 90
Query 90 FKRFMGEADDAPWMPTKTYKVPKIIIDNEEGGAVRAYPDESTILDYMGVPPKAIPVGGTG 149
++ MGE + W P Y VP+I EGG + TI DYMG+P G
Sbjct 91 WRELMGENTQSAWTPQVEYSVPQITA--PEGGW-----NVGTIADYMGIP------TGVS 137
Query 150 RIEINALPVRAYVKIWNEFFRDQNVGNPAVLNTGDEDEAYRAGSGNESEVSEEKILEYAH 209
+ +NA+P RAY I NE+FRD+N+ +P + GD A AG + V++
Sbjct 138 GLSVNAMPFRAYALICNEWFRDENLTDPLNIPVGD---ATVAGVNTGTYVTD------VA 188
Query 210 IGGYCLPVNRFHDYFSSCLPYPQRGPEVTIALTGN--APLRAYSEKD--LNNRKIGTGFF 265
GG ++HDYF+SCLP PQ+GP+V I+ G+ P+ A + LN G F
Sbjct 189 KGGLPFKAAKYHDYFTSCLPAPQKGPDVLISAVGSGIVPVTATDNDNDSLNVNSPGMRFV 248
Query 266 NNEYNTGIVNHTNISFTKEGTKFSVNKNNNGNTAPLVNGQYIQTMSQDDANFF-DAWLGT 324
N + VN+ ++F G + V +T I +S N + D T
Sbjct 249 GNSSTS--VNY--LAF-GGGDGYVVTDTPKPSTP-------IHGISMIPTNLWADLSTAT 296
Query 325 DLSNIEAATINQLRQAFAVQHYYEALARGGSRYREQVRALFGVSISDKTVQIPEYLGGGR 384
DL ATINQLR AF +Q YE ARGG+RY E +++ FGV+ D +Q PEYLGG R
Sbjct 297 DL---PVATINQLRTAFQIQKLYERDARGGTRYIEILKSHFGVTSPDARLQRPEYLGGSR 353
Query 385 YHVNMNQIVQTSGQESNYGTPIGETGAMSVTPINESSFTKSFEEHGFVIGVMCVRHDHSY 444
+N+NQ++Q+S TP G A S+T + S FTKSF EHGF+IG+M R+DHSY
Sbjct 354 VPININQVIQSS---ETGATPQGNAAAYSLTTDSHSEFTKSFVEHGFIIGLMVARYDHSY 410
Query 445 QQGLERFWSRSDRLDYYFPQFANLGEQPVKKKEIMLTGKSTDEETFGYQEAWADYRMKPN 504
QQGL+RFWSR DR DYY+P FANLGE VK KEI G D+E FGYQEAWADYR KP+
Sbjct 411 QQGLQRFWSRKDRFDYYWPVFANLGEMAVKNKEIFAQGTDVDDEVFGYQEAWADYRYKPS 470
Query 505 RVSGKMRSNAEGTLDFWHYADNYATVPTLSQEWMKEGKNEIARTLIVEN--EPQFFGAIR 562
V+G+MRS +LD WH AD+Y +P+LS W++E + + R L V + Q F I
Sbjct 471 VVTGEMRSQYAQSLDIWHLADDYENLPSLSDSWIREDSSTVNRVLAVSDSVSAQLFCDIY 530
Query 563 VMNKTTRCMPLYSVPGL 579
+ TR MPLYS+PGL
Sbjct 531 IRCLATRPMPLYSIPGL 547
>gi|575094415|emb|CDL65790.1| unnamed protein product [uncultured bacterium]
Length=569
Score = 434 bits (1116), Expect = 1e-141, Method: Compositional matrix adjust.
Identities = 240/609 (39%), Positives = 340/609 (56%), Gaps = 67/609 (11%)
Query 1 MNRNNERHFNQVPETHVSRTRFKRDQNILTTFDSGKLIPFYVDEVLPGDTFSVDTAAIIR 60
MNRN E H++Q+P ++ R +FKRD + LTT + G L+P YVDEVLPGDT + +++R
Sbjct 1 MNRNAEAHYSQIPHANIQRAKFKRDFSYLTTINEGDLVPIYVDEVLPGDTIKIKQRSLVR 60
Query 61 MTTPKYPVMDDAYIDFYYFYCPNRILWDNFKRFMGEADDAPWMPTKTYKVPKIIIDNEEG 120
M+TP YPVMD+ Y+D +YF+ P R++WD+++ MGE + W P Y P + G
Sbjct 61 MSTPLYPVMDNCYLDIWYFFVPCRLVWDHWQNLMGENTKSYWAPDVQYTTP--LTSAPSG 118
Query 121 GAVRAYPDESTILDYMGVPPKAIPVGGTGRIEINALPVRAYVKIWNEFFRDQNVGNPAVL 180
G TI DYMG+P G I++N++P+RAY +IWNE+FRD+N+ P
Sbjct 119 GW-----QVGTIADYMGIPT------GVSGIKVNSMPMRAYARIWNEWFRDENLQQPV-- 165
Query 181 NTGDEDEAYRAGSGNESEVSEEKILEYAHIGGYCLPVNRFHDYFSSCLPYPQRGPEVTIA 240
T D+A GS +E+++ A GG L V +F DYF+SCLP PQ+G +
Sbjct 166 -TQHSDDATTTGSNTGTELTD------AESGGLPLKVAKFKDYFTSCLPAPQKGEAIGFD 218
Query 241 LTGNAPLRA------------YSEKDLNNRKIGTGFFNNEYNTGIVNHTNISFTKEGTKF 288
++ ++ D+ R+ YNT N +I+ T+
Sbjct 219 FNQTPKVKGIGLVFPLETNTGHTATDILWRQPDAQLVGENYNTSYNNFNSIT-----TQT 273
Query 289 SVNKN-----NNGNTAPLVNGQYIQTMSQDDANFFDAWLGTDLSNIEAA--------TIN 335
+VN NNG P+++ ++ +DD N G + + A +IN
Sbjct 274 TVNGKKAFFFNNGK-GPMLSARF-----EDDYNG-----GVEQVELTAVAENSTNFLSIN 322
Query 336 QLRQAFAVQHYYEALARGGSRYREQVRALFGVSISDKTVQIPEYLGGGRYHVNMNQIVQT 395
LRQA A+QH EA ARGG+RY E ++ FGVS D +Q EY+GG R +N++Q++Q+
Sbjct 323 DLRQAIALQHILEADARGGTRYVEILKNEFGVSSPDARLQRSEYIGGERIPINVSQVIQS 382
Query 396 SGQESNYGTPIGETGAMSVTPINESSFTKSFEEHGFVIGVMCVRHDHSYQQGLERFWSRS 455
S ++ +P G A S+T + S EHG+++G+ +R DHSYQQGL R W+RS
Sbjct 383 SASDTT--SPQGNAAAYSLTTSANTIRAYSAVEHGYILGLAAIRVDHSYQQGLSRMWTRS 440
Query 456 DRLDYYFPQFANLGEQPVKKKEIMLTGKSTDEETFGYQEAWADYRMKPNRVSGKMRSNAE 515
DR YY P ANLGEQ V +EI G + D E FGYQEAWADYR + N ++G+MRS
Sbjct 441 DRFSYYHPMLANLGEQAVLNQEIYAQGTTADTEVFGYQEAWADYRYRTNMITGEMRSTYA 500
Query 516 GTLDFWHYADNYATVPTLSQEWMKEGKNEIARTLIV--ENEPQFFGAIRVMNKTTRCMPL 573
+LD WHY D Y +P LS +W+KEG+ I RTL V EN QF + R MP+
Sbjct 501 QSLDAWHYGDKYTDLPRLSNDWIKEGQENIDRTLAVQSENSHQFICNLYFDQTWVRPMPI 560
Query 574 YSVPGLEKL 582
YSVPGL +
Sbjct 561 YSVPGLSMI 569
>gi|575094496|emb|CDL65862.1| unnamed protein product [uncultured bacterium]
Length=568
Score = 421 bits (1081), Expect = 2e-136, Method: Compositional matrix adjust.
Identities = 239/593 (40%), Positives = 335/593 (56%), Gaps = 46/593 (8%)
Query 3 RNNERHFNQVPET-HVSRTRFKRDQNILTTFDSGKLIPFYVDEVLPGDTFSVDTAAIIRM 61
RN F++ P T + R+ F R T+ + G+LIPFY DEVLPGDTF V T ++R+
Sbjct 2 RNENSRFSENPVTLDIQRSTFNRSSTYKTSANIGELIPFYYDEVLPGDTFQVKTNKVVRL 61
Query 62 TTPKYPVMDDAYIDFYYFYCPNRILWDNFKRFMGEADDAPWMPTKTYKVPKIIIDNEEGG 121
MD+ Y D YYF+ PNR++W++++ FMGE W+P Y +P+I G
Sbjct 62 QPLVSAPMDNLYFDTYYFFVPNRLVWEHWEEFMGENKQGAWIPQTEYTIPQITSPASTGF 121
Query 122 AVRAYPDESTILDYMGVPPKAIPVGGTGRIEINALPVRAYVKIWNEFFRDQNVGNPAVLN 181
+ TI DY G+P G + ++ALP RAY I +E+FRDQN+ P LN
Sbjct 122 EI------GTIADYFGIP------TGVPNLSVSALPFRAYALIVDEWFRDQNLQLP--LN 167
Query 182 TGDEDEAYRAGSGNESEVSEEKILEYAHIGGYCLPVNRFHDYFSSCLPYPQRGPEVTIAL 241
+D + + + K GG ++HDYF+SCLP PQ+GP+VTIA
Sbjct 168 IPLDDTTLQGVNTGDYVTDTVK-------GGKPFVAAKYHDYFTSCLPSPQKGPDVTIAA 220
Query 242 TGNAPLRAYSEKDLNNRKIGTGFFNNEYNTGIVNHTNISFTKEGTKF-SVNKNNNGNTAP 300
G+ P+ Y+ NN Y ++ ++SF++ SV + + P
Sbjct 221 VGDFPV--YTGDPHNNNGSNKAL---HYGISNISSGSVSFSQGNYIIPSVLTTGSTQSVP 275
Query 301 L---VNGQYIQTMSQDDANFFDAWLGTDLS---------NIEAATINQLRQAFAVQHYYE 348
+N I TM+ + D+ G+ LS + A TINQLR AF +Q YE
Sbjct 276 AQGKLNASNI-TMTTSPGSP-DSSFGSKLSVYPDNLYASSGTATTINQLRMAFQIQKLYE 333
Query 349 ALARGGSRYREQVRALFGVSISDKTVQIPEYLGGGRYHVNMNQIVQTSGQESNYGTPIGE 408
AR GSRYRE +R+ F V+ D +Q+PEYLGG R +N+NQ+VQTS +++ +P G
Sbjct 334 KDARAGSRYRELIRSHFSVTPLDARMQVPEYLGGNRIPININQVVQTS--QTSDVSPQGN 391
Query 409 TGAMSVTPINESSFTKSFEEHGFVIGVMCVRHDHSYQQGLERFWSRSDRLDYYFPQFANL 468
S+T + F KSF EHG +IGV R+DH+YQQG+ + WSR R DYY+P AN+
Sbjct 392 VAGQSLTSDSHGDFIKSFTEHGMLIGVAVARYDHTYQQGVSKLWSRKTRFDYYWPVLANI 451
Query 469 GEQPVKKKEIMLTGKSTDEETFGYQEAWADYRMKPNRVSGKMRSNAEGTLDFWHYADNYA 528
GEQ V KEI G + DEE FGYQEAWA+YR KP+ V+G+MRS+A +LD WH+AD+Y
Sbjct 452 GEQAVLNKEIYAQGTAQDEEVFGYQEAWAEYRYKPSIVTGEMRSSARTSLDSWHFADDYN 511
Query 529 TVPTLSQEWMKEGKNEIARTLIVENEP--QFFGAIRVMNKTTRCMPLYSVPGL 579
++P LS +W+KE K I R L V + Q+F + N+TTR +P YS+PGL
Sbjct 512 SLPKLSADWIKEDKTNIDRVLAVSSSVSNQYFADFYIENETTRALPFYSIPGL 564
>gi|557745632|ref|YP_008798242.1| major capsid protein [Marine gokushovirus]
gi|530695345|gb|AGT39902.1| major capsid protein [Marine gokushovirus]
Length=538
Score = 394 bits (1012), Expect = 2e-126, Method: Compositional matrix adjust.
Identities = 233/591 (39%), Positives = 318/591 (54%), Gaps = 92/591 (16%)
Query 6 ERHFNQVPETHVSRTRFKRDQNILTTFDSGKLIPFYVDEVLPGDTFSVDTAAIIRMTTPK 65
+ F++VP + R+ F R + TTF++G+L+P YVDE LPGDTFS + A R+ TP
Sbjct 18 QHQFSEVPHADIQRSTFDRSHGLKTTFNAGQLVPIYVDEALPGDTFSCNLTAFSRLATPI 77
Query 66 YPVMDDAYIDFYYFYCPNRILWDNFKRFMGEADDAPWMPTKTYK---------------- 109
+P MD+A++D ++F P R++WD+F+ FMGE TKTYK
Sbjct 78 HPTMDNAFMDTHFFAVPVRLVWDDFEEFMGE--------TKTYKAAGSDRLDGTPDFSVA 129
Query 110 --VPKIIIDNEEGGAVRAYPDESTILDYMGVPPKAIPVGGTGRIEINALPVRAYVKIWNE 167
VP I + G A E+++ DY G+P K VGG +E +AL RAY +WN+
Sbjct 130 APVPPTITASGSGEA------EASLSDYFGIPTK---VGG---LEFSALWHRAYTLVWND 177
Query 168 FFRDQNVGNPAVLNTGDEDEAYRAGSGNESEVSEEKILEYAHIGGYCLPVNRFHDYFSSC 227
+FRD+N+ P ++T SGN++ YA L + HDYF+S
Sbjct 178 WFRDENLQAPKTIDTT---------SGNDTTT-------YA-----LLNRGKKHDYFTSA 216
Query 228 LPYPQRGPEVTIALTGNAPLRAYSEKDLNNRKIGTGFFNNEYNTGIVNHTNISFTKEGTK 287
LP+PQ+G +VTI L +AP+ N +N T
Sbjct 217 LPWPQKGADVTIPLGTSAPVTT------------------------ANSSNQDVT----- 247
Query 288 FSVNKNNNGNTAPLVNGQYIQTMSQDDANFFDAWLGTDLSNIEAATINQLRQAFAVQHYY 347
+ N GNT +N D+ L DLS +ATINQLR AFA Q +
Sbjct 248 --IFTPNIGNTHRFLNSASTNVYPGDENTDEARRLYADLSEATSATINQLRLAFATQKFL 305
Query 348 EALARGGSRYREQVRALFGVSISDKTVQIPEYLGGGRYHVNMNQIVQTSGQESNYGTPIG 407
E ARGGSRY E ++ F V+ D +Q PEYLGGG VN++ + QTS ++ TP G
Sbjct 306 EIQARGGSRYIEVIKNHFNVTSPDARLQRPEYLGGGSSPVNISPVAQTSSTDAT--TPQG 363
Query 408 ETGAMSVTPINESSFTKSFEEHGFVIGVMCVRHDHSYQQGLERFWSRSDRLDYYFPQFAN 467
A+ T ++ SFTKSF EH VIG++ VR D +YQQGL R +SR DYY+P +
Sbjct 364 NLSAIGTTVLSGHSFTKSFTEHTIVIGMVSVRTDLTYQQGLNRMFSRETIYDYYWPTLST 423
Query 468 LGEQPVKKKEIMLTGKSTDEETFGYQEAWADYRMKPNRVSGKMRSNAEGTLDFWHYADNY 527
+GEQ VK KEI G + DE TFGYQE +A+YR KP+ V+GK RSNA GTL+ WHYA Y
Sbjct 424 IGEQAVKNKEIYAQGSAADETTFGYQERYAEYRYKPSSVTGKFRSNATGTLESWHYAQEY 483
Query 528 ATVPTLSQEWMKEGKNEIARTLIVENEPQFFGAIRVMNKTTRCMPLYSVPG 578
A++P L W++ + RTL V +EPQF + TR MP+ S+PG
Sbjct 484 ASLPLLGDSWIQVTDTNVQRTLAVASEPQFIFDSLFKLRCTRPMPVNSIPG 534
>gi|530695351|gb|AGT39907.1| major capsid protein [Marine gokushovirus]
Length=539
Score = 389 bits (999), Expect = 2e-124, Method: Compositional matrix adjust.
Identities = 241/590 (41%), Positives = 333/590 (56%), Gaps = 67/590 (11%)
Query 1 MNRN---NERHFNQVPETHVSRTRFKRDQNILTTFDSGKLIPFYVDEVLPGDTFSVDTAA 57
M+RN + F+ +P + R++F + + T FDSG L+P VDEVLPGD+ ++ A
Sbjct 2 MHRNKSASAHQFSMIPRAEIPRSKFDAQKTLKTAFDSGYLVPILVDEVLPGDSMNLRMTA 61
Query 58 IIRMTTPKYPVMDDAYIDFYYFYCPNRILWDNFKRFMGEADDAPWMPTKTYKVPKIIIDN 117
R+ TP +PVMD+ Y+D ++F+ PNR+LW N++RFMGE D P + Y +P + N
Sbjct 62 FTRLATPLFPVMDNMYLDTFFFFVPNRLLWSNWQRFMGERDPDP-DSSIDYTIPTMTSPN 120
Query 118 EEGGAVRAYPDESTILDYMGVPPKAIPVGGTGRIEINALPVRAYVKIWNEFFRDQNVGNP 177
G AV +++ DYMG+P A V I N+L RAY IWNE+FRD+N+ +
Sbjct 121 G-GYAV------NSLQDYMGLP-TAGQVDAGSSISHNSLFTRAYNLIWNEWFRDENLQDS 172
Query 178 AVLNTGDEDEAYRAGSGNESEVSEEKILEYAHIGGYCLPVNRFHDYFSSCLPYPQRGPEV 237
V++ GD + Y +Y L + HDYF+S LP+PQ+G V
Sbjct 173 VVVDKGDGPDTYT---------------DYT-----LLRRGKRHDYFTSALPWPQKGDAV 212
Query 238 TIALTGNAPLRAYSEKDLNNRKIGTGFFNNEYNTGIVNHTNISFTKEGTKFSVNKNNNGN 297
T+ L G+A ++ G + E +TG V T ++ SV+K NGN
Sbjct 213 TLPLGGSA--------NVVYNDTGDPAYIREVSTGNVWTTP-------SRESVSKEANGN 257
Query 298 -TAPL--VNGQYIQTMSQDDANFFDAWLGTDLSNIEAATINQLRQAFAVQHYYEALARGG 354
+ P VN QY D N L DLS AATIN +RQ+F +Q E ARGG
Sbjct 258 MSVPTGSVNAQY-------DPN---GSLVADLSTATAATINAIRQSFQIQRLLERDARGG 307
Query 355 SRYREQVRALFGVSISDKTVQIPEYLGGGRYHVNMNQIVQTSGQ-ESNYGTPIGETGAMS 413
+RY E VR+ FGV D +Q PEYLGGG + +N + Q S S TP+G GA+
Sbjct 308 TRYTEIVRSHFGVISPDARMQRPEYLGGGSAPIIVNPVAQQSASGASGTDTPLGTLGAVG 367
Query 414 VTPINESSFTKSFEEHGFVIGVMCVRHDHSYQQGLERFWSRSDRLDYYFPQFANLGEQPV 473
+ F SF EHG V+G+ VR D +YQQGL R +SRS R D++FP F++LGEQP+
Sbjct 368 TGLASGHGFASSFTEHGVVVGLCSVRADLTYQQGLHRMFSRSTRYDFFFPVFSHLGEQPI 427
Query 474 KKKEIMLTGKSTDEETFGYQEAWADYRMKPNRVSGKMRSNAEGTLDFWHYADNYATVPTL 533
KE+ TG STD++ FGYQEAWA+YR KP++V+G MRS A GTLD WH A N+ ++PTL
Sbjct 428 LNKELYATGTSTDDDVFGYQEAWAEYRYKPSQVTGLMRSTAAGTLDAWHLAQNFGSLPTL 487
Query 534 SQEWMKEGKNEIARTLIVENEP---QF-FGAIRVMNKTTRCMPLYSVPGL 579
+ ++ E + R + V +E QF F A +N R MP+YSVPGL
Sbjct 488 NSTFI-EDTPPVDRVVAVGSEANGQQFIFDAFFDINM-ARPMPMYSVPGL 535
>gi|313766927|gb|ADR80653.1| putative major coat protein [Uncultured Microviridae]
Length=533
Score = 384 bits (985), Expect = 1e-122, Method: Compositional matrix adjust.
Identities = 225/574 (39%), Positives = 317/574 (55%), Gaps = 79/574 (14%)
Query 9 FNQVPETHVSRTRFKRDQNILTTFDSGKLIPFYVDEVLPGDTFSVDTAAIIRMTTPKYPV 68
F++VP+ + R+ F R + TTF+SG LIP YVDEVLPGDTF ++ R+ TP YPV
Sbjct 17 FSRVPQADIQRSTFSRVHGLKTTFNSGDLIPIYVDEVLPGDTFQMNATGFGRLATPLYPV 76
Query 69 MDDAYIDFYYFYCPNRILWDNFKRFMGEADDAPWMPTKTYKVPKIIIDNEEGGAVRAYPD 128
MD+ Y++ ++FY PNRI+WDN+++F G DD + + VP+I A
Sbjct 77 MDNMYVETFFFYVPNRIIWDNWEKFNGAQDDP--NDSTDFLVPQI---------QSATVA 125
Query 129 ESTILDYMGVPPKAIPVGGTGRIEINALPVRAYVKIWNEFFRDQNVGNPAVLNTGDEDEA 188
E ++ DYMG+P + + G I+ N L RAY IWNE+FRD+N+ + + D +
Sbjct 126 EGSLFDYMGLPTQ---IAG---IDFNNLHGRAYNLIWNEWFRDENLQDSLGVPKDDGPDT 179
Query 189 YRAGSGNESEVSEEKILEYAHIGGYCL-PVNRFHDYFSSCLPYPQRGPEVTIALTGNAPL 247
Y GY + + HDYF+S LP+PQ+G V++ L +A +
Sbjct 180 YT---------------------GYTIQKRGKRHDYFTSALPWPQKGDAVSLPLGTSADI 218
Query 248 R--AYSEKDLNNRKIGTGFFNNEYNTGIVNHTNISFTKEGTKFSVNKNNNGNTAPLVNGQ 305
A + D+ +G+ F + T V +G T P N
Sbjct 219 HTAAAAGTDIGIYSVGSSDF-----------------RLLTSDPVEVALSGGTPPETNKM 261
Query 306 YIQTMSQDDANFFDAWLGTDLSNIEAATINQLRQAFAVQHYYEALARGGSRYREQVRALF 365
+ DLSN AATINQLR+AF +Q YE ARGG+RY E +++ F
Sbjct 262 F-----------------ADLSNATAATINQLREAFQIQRLYEKDARGGTRYTEILQSHF 304
Query 366 GVSISDKTVQIPEYLGGGRYHVNMNQIVQTSGQESNYGTPIGETGAMSVTPINESSFTKS 425
GV+ D +Q PEYLGG + V M + QTS +S +P G A+ T + F+KS
Sbjct 305 GVTSPDARLQRPEYLGGQKTEVMMQTVPQTSSTDST--SPQGNLAALG-TATSRGGFSKS 361
Query 426 FEEHGFVIGVMCVRHDHSYQQGLERFWSRSDRLDYYFPQFANLGEQPVKKKEIMLTGKST 485
F EHG +IG+ CV D +YQQG+ R WSR DR D+Y+P A+LGEQ V +EI G S
Sbjct 362 FVEHGVLIGLACVFADLTYQQGMNRMWSRRDRWDFYWPSLAHLGEQAVLNQEIYTQGTSA 421
Query 486 DEETFGYQEAWADYRMKPNRVSGKMRSNAEGTLDFWHYADNYATVPTLSQEWMKEGKNEI 545
D +TFGYQE +A+YR KP++++GKMRSNA GTLD WH A ++ +P L+ +++E +
Sbjct 422 DTQTFGYQERFAEYRYKPSQITGKMRSNATGTLDAWHLAQDFTALPALNASFIEENP-PV 480
Query 546 ARTLIVENEPQFFGAIRVMNKTTRCMPLYSVPGL 579
R + V +EP+F KTTR MP+YSVPGL
Sbjct 481 DRVIAVPSEPEFIWDWYFDLKTTRPMPVYSVPGL 514
Lambda K H a alpha
0.317 0.135 0.410 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 4286665841916