bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: All non-redundant GenBank CDS translations+PDB+SwissProt+PIR+PRF
excluding environmental samples from WGS projects
49,011,213 sequences; 17,563,301,199 total letters
Query= Contig-20_CDS_annotation_glimmer3.pl_2_3
Length=101
Score E
Sequences producing significant alignments: (Bits) Value
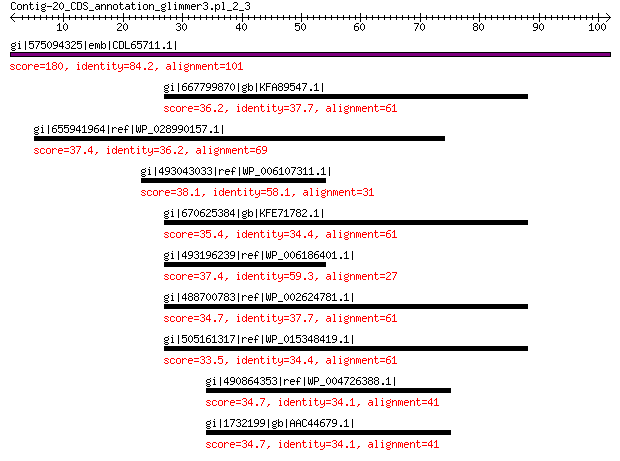
gi|575094325|emb|CDL65711.1| unnamed protein product 180 5e-56
gi|667799870|gb|KFA89547.1| flagellar biosynthesis protein FlhB 36.2 0.49
gi|655941964|ref|WP_028990157.1| hypothetical protein 37.4 0.52
gi|493043033|ref|WP_006107311.1| histidine ammonia-lyase 38.1 0.58
gi|670625384|gb|KFE71782.1| Flagellar biosynthesis protein FlhB 35.4 0.99
gi|493196239|ref|WP_006186401.1| histidine ammonia-lyase 37.4 1.00
gi|488700783|ref|WP_002624781.1| Flagellar biosynthesis protein ... 34.7 1.9
gi|505161317|ref|WP_015348419.1| FlhB/HrpN/YscU/SpaS family protein 33.5 4.5
gi|490864353|ref|WP_004726388.1| PTS N-acetylgalactosamine trans... 34.7 5.2
gi|1732199|gb|AAC44679.1| PTS permease for mannose subunit IIIMa... 34.7 5.2
>gi|575094325|emb|CDL65711.1| unnamed protein product [uncultured bacterium]
Length=107
Score = 180 bits (456), Expect = 5e-56, Method: Compositional matrix adjust.
Identities = 85/101 (84%), Positives = 96/101 (95%), Gaps = 0/101 (0%)
Query 1 MKVLSRINPSEPFYRVSVKRVSTDEPVVSGLSITPSDIERLARQGVPVSVPNANSFYSID 60
+K+LSRINP E ++RV+ +R +DEPVVSGL+ITPSDIE+LARQGVPVSVPNANSFYSID
Sbjct 7 VKILSRINPREVYHRVTCRRTISDEPVVSGLAITPSDIEKLARQGVPVSVPNANSFYSID 66
Query 61 SGLEVPPELKVDADRNSLWEMSQQSKARIMKARKREKDHLT 101
SGLEVPPELKVDADRN+LWE+SQQSKARIMKARKREKDHLT
Sbjct 67 SGLEVPPELKVDADRNTLWELSQQSKARIMKARKREKDHLT 107
>gi|667799870|gb|KFA89547.1| flagellar biosynthesis protein FlhB [Cystobacter violaceus Cb
vi76]
Length=93
Score = 36.2 bits (82), Expect = 0.49, Method: Compositional matrix adjust.
Identities = 23/66 (35%), Positives = 36/66 (55%), Gaps = 5/66 (8%)
Query 27 VVSGLSITPSDIERLARQ-GVPV--SVPNANSFYSIDSGLEVPPEL--KVDADRNSLWEM 81
V G+ + I +ARQ +PV +V AN+ Y ID G E+P EL V N ++E+
Sbjct 24 VAKGMRLKAEKIREIARQYNIPVMRNVSLANALYRIDVGQEIPEELYDAVAEVLNFIFEL 83
Query 82 SQQSKA 87
++ +A
Sbjct 84 QREQQA 89
>gi|655941964|ref|WP_028990157.1| hypothetical protein [Thermithiobacillus tepidarius]
Length=156
Score = 37.4 bits (85), Expect = 0.52, Method: Compositional matrix adjust.
Identities = 25/69 (36%), Positives = 39/69 (57%), Gaps = 8/69 (12%)
Query 5 SRINPSEPFYRVSVKRVSTDEPVVSGLSITPSDIERLARQGVPVSVPNANSFYSIDSGLE 64
S INP+ P + V+ ++V D + I+P DI + R G+PV+V A S +DSG+
Sbjct 83 SPINPNHPAF-VATEQVRDDGQTM----ISPVDINKAYRDGLPVTVTPARSLKKVDSGIT 137
Query 65 VPPELKVDA 73
+L +DA
Sbjct 138 ---QLSIDA 143
>gi|493043033|ref|WP_006107311.1| histidine ammonia-lyase [Natrialba asiatica]
gi|445652676|gb|ELZ05562.1| histidine ammonia-lyase [Natrialba asiatica DSM 12278]
Length=524
Score = 38.1 bits (87), Expect = 0.58, Method: Composition-based stats.
Identities = 18/32 (56%), Positives = 25/32 (78%), Gaps = 1/32 (3%)
Query 23 TDEPVV-SGLSITPSDIERLARQGVPVSVPNA 53
TD+PVV G ++TP+ +ER+AR G PVSVP +
Sbjct 2 TDDPVVVDGETLTPAAVERVARAGAPVSVPES 33
>gi|670625384|gb|KFE71782.1| Flagellar biosynthesis protein FlhB [Hyalangium minutum]
gi|670631701|gb|KFE77751.1| Flagellar biosynthesis protein FlhB [Enhygromyxa salina]
Length=94
Score = 35.4 bits (80), Expect = 0.99, Method: Compositional matrix adjust.
Identities = 21/66 (32%), Positives = 36/66 (55%), Gaps = 5/66 (8%)
Query 27 VVSGLSITPSDIERLARQ-GVPV--SVPNANSFYSIDSGLEVPPEL--KVDADRNSLWEM 81
V G+ + I +A+Q +P+ ++P AN+ Y +D G EVP EL V N ++ +
Sbjct 24 VAKGMRLKAEKIREIAKQYNIPIMKNLPLANALYRVDVGQEVPEELYDAVAEVLNFVYAL 83
Query 82 SQQSKA 87
Q+ +A
Sbjct 84 QQEQQA 89
>gi|493196239|ref|WP_006186401.1| histidine ammonia-lyase [Natrinema pallidum]
gi|445621345|gb|ELY74821.1| histidine ammonia-lyase [Natrinema pallidum DSM 3751]
Length=524
Score = 37.4 bits (85), Expect = 1.00, Method: Composition-based stats.
Identities = 16/27 (59%), Positives = 22/27 (81%), Gaps = 0/27 (0%)
Query 27 VVSGLSITPSDIERLARQGVPVSVPNA 53
VV G S+TP+D+ER+AR+G VSVP +
Sbjct 7 VVDGTSLTPADVERVAREGATVSVPES 33
>gi|488700783|ref|WP_002624781.1| Flagellar biosynthesis protein FlhB [Cystobacter fuscus]
gi|528053281|gb|EPX58088.1| Flagellar biosynthesis protein FlhB [Cystobacter fuscus DSM 2262]
Length=93
Score = 34.7 bits (78), Expect = 1.9, Method: Compositional matrix adjust.
Identities = 23/66 (35%), Positives = 35/66 (53%), Gaps = 5/66 (8%)
Query 27 VVSGLSITPSDIERLARQG-VPV--SVPNANSFYSIDSGLEVPPEL--KVDADRNSLWEM 81
V GL + I +ARQ +P+ +V AN+ Y +D G EVP EL V N ++E+
Sbjct 24 VAKGLRLKAEKIREIARQNNIPLMKNVALANALYRVDVGQEVPEELYDAVAEILNFVYEL 83
Query 82 SQQSKA 87
+ +A
Sbjct 84 QRAGQA 89
>gi|505161317|ref|WP_015348419.1| FlhB/HrpN/YscU/SpaS family protein [Myxococcus stipitatus]
gi|442319821|ref|YP_007359842.1| FlhB/HrpN/YscU/SpaS family protein [Myxococcus stipitatus DSM
14675]
gi|441487463|gb|AGC44158.1| FlhB/HrpN/YscU/SpaS family protein [Myxococcus stipitatus DSM
14675]
Length=93
Score = 33.5 bits (75), Expect = 4.5, Method: Compositional matrix adjust.
Identities = 21/66 (32%), Positives = 36/66 (55%), Gaps = 5/66 (8%)
Query 27 VVSGLSITPSDIERLARQ-GVPV--SVPNANSFYSIDSGLEVPPEL--KVDADRNSLWEM 81
V GL + I +A++ +P+ +VP AN+ Y ++ G EVP EL V N ++E+
Sbjct 24 VAKGLRLKAEKIRAIAKEHNIPIMRNVPLANALYRVEVGQEVPEELYDAVAEVLNFIYEL 83
Query 82 SQQSKA 87
++ A
Sbjct 84 QREHAA 89
>gi|490864353|ref|WP_004726388.1| PTS N-acetylgalactosamine transporter subunit IIB [Vibrio furnissii]
gi|375131373|ref|YP_004993473.1| PTS permease for mannose subunit IIIMan C [Vibrio furnissii NCTC
11218]
gi|260616874|gb|EEX42059.1| PTS system N-acetylgalactosamine-specific IIB component [Vibrio
furnissii CIP 102972]
gi|315180547|gb|ADT87461.1| PTS permease for mannose subunit IIIMan C [Vibrio furnissii NCTC
11218]
Length=157
Score = 34.7 bits (78), Expect = 5.2, Method: Compositional matrix adjust.
Identities = 14/41 (34%), Positives = 22/41 (54%), Gaps = 0/41 (0%)
Query 34 TPSDIERLARQGVPVSVPNANSFYSIDSGLEVPPELKVDAD 74
TP D RL GVP++ N + + ID ++ + VDA+
Sbjct 85 TPHDFRRLVEGGVPIAAINVGNMHYIDGKTQISKTVSVDAE 125
>gi|1732199|gb|AAC44679.1| PTS permease for mannose subunit IIIMan C terminal domain [Vibrio
furnissii]
Length=157
Score = 34.7 bits (78), Expect = 5.2, Method: Compositional matrix adjust.
Identities = 14/41 (34%), Positives = 22/41 (54%), Gaps = 0/41 (0%)
Query 34 TPSDIERLARQGVPVSVPNANSFYSIDSGLEVPPELKVDAD 74
TP D RL GVP++ N + + ID ++ + VDA+
Sbjct 85 TPHDFRRLVEGGVPIAAINVGNMHYIDGKTQISKTVSVDAE 125
Lambda K H a alpha
0.312 0.128 0.355 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 438108004959