bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: All non-redundant GenBank CDS translations+PDB+SwissProt+PIR+PRF
excluding environmental samples from WGS projects
49,011,213 sequences; 17,563,301,199 total letters
Query= Contig-21_CDS_annotation_glimmer3.pl_2_3
Length=101
Score E
Sequences producing significant alignments: (Bits) Value
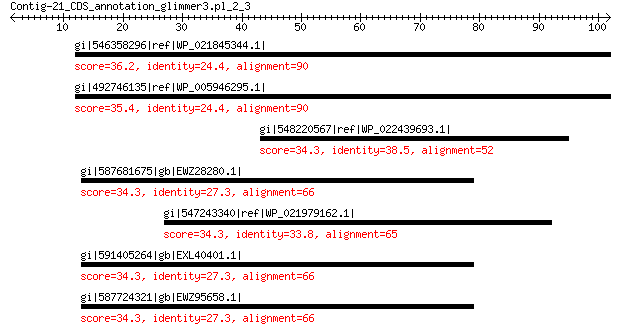
gi|546358296|ref|WP_021845344.1| putative uncharacterized protein 36.2 2.7
gi|492746135|ref|WP_005946295.1| hypothetical protein 35.4 3.8
gi|548220567|ref|WP_022439693.1| fe2+/Zn2+ uptake regulation pro... 34.3 5.9
gi|587681675|gb|EWZ28280.1| beta-glucosidase 34.3 9.1
gi|547243340|ref|WP_021979162.1| beta-N-acetylglucosaminidase 34.3 9.6
gi|591405264|gb|EXL40401.1| beta-glucosidase 34.3 9.8
gi|587724321|gb|EWZ95658.1| beta-glucosidase 34.3 9.9
>gi|546358296|ref|WP_021845344.1| putative uncharacterized protein [Blautia hydrogenotrophica CAG:147]
gi|524855697|emb|CCX58499.1| putative uncharacterized protein [Blautia hydrogenotrophica CAG:147]
Length=2797
Score = 36.2 bits (82), Expect = 2.7, Method: Composition-based stats.
Identities = 22/90 (24%), Positives = 44/90 (49%), Gaps = 0/90 (0%)
Query 12 ESGKEEEEIKDVNLEIEEREVSENGPFVLIRNKKNKWVITTCGALVNGKEFDTKEDAEKH 71
E KE+ +I+ N E+ + +G + I + W +T+ G K F+ KE+A+K+
Sbjct 920 EDAKEKADIQVKNAGGEKIYPNSDGSYSFIEDGTYNWTVTSEGYWTESKTFEVKEEADKN 979
Query 72 LAKKEWDDILTAALIFFTHVKNQMENTQKE 101
+ +E ++ + F V ++ +N E
Sbjct 980 VEFREALEMSPTYPVKFEFVSDKPQNQTIE 1009
>gi|492746135|ref|WP_005946295.1| hypothetical protein, partial [Blautia hydrogenotrophica]
gi|225040024|gb|EEG50270.1| hypothetical protein RUMHYD_00803, partial [Blautia hydrogenotrophica
DSM 10507]
Length=705
Score = 35.4 bits (80), Expect = 3.8, Method: Composition-based stats.
Identities = 22/90 (24%), Positives = 44/90 (49%), Gaps = 0/90 (0%)
Query 12 ESGKEEEEIKDVNLEIEEREVSENGPFVLIRNKKNKWVITTCGALVNGKEFDTKEDAEKH 71
E KE+ +I+ N E+ + +G + I + W +T+ G K F+ KE+A+K+
Sbjct 371 EDAKEKADIQVKNAGGEKIYPNSDGSYSFIEDGTYNWTVTSEGYWTESKTFEVKEEADKN 430
Query 72 LAKKEWDDILTAALIFFTHVKNQMENTQKE 101
+ +E ++ + F V ++ +N E
Sbjct 431 VEFREALEMSPTYPVKFEFVSDKPQNQTIE 460
>gi|548220567|ref|WP_022439693.1| fe2+/Zn2+ uptake regulation proteins [Clostridium sp. CAG:411]
gi|524742633|emb|CDE43344.1| fe2+/Zn2+ uptake regulation proteins [Clostridium sp. CAG:411]
Length=149
Score = 34.3 bits (77), Expect = 5.9, Method: Compositional matrix adjust.
Identities = 20/55 (36%), Positives = 31/55 (56%), Gaps = 4/55 (7%)
Query 43 NKKNKWVITTCGALVNGKE---FDTKEDAEKHLAKKEWDDILTAALIFFTHVKNQ 94
N+KN + I+ C + KE + ++ HL+ KEW D+L A L + +VKNQ
Sbjct 83 NRKNMYKIS-CSMDCDKKEACQIELDDNTVCHLSPKEWKDVLRAGLEQYGYVKNQ 136
>gi|587681675|gb|EWZ28280.1| beta-glucosidase [Fusarium oxysporum Fo47]
Length=842
Score = 34.3 bits (77), Expect = 9.1, Method: Composition-based stats.
Identities = 18/68 (26%), Positives = 33/68 (49%), Gaps = 2/68 (3%)
Query 13 SGKEEEEIKDVNLEIEEREVSEN--GPFVLIRNKKNKWVITTCGALVNGKEFDTKEDAEK 70
+ ++E + VN E+ +R + E PF ++ N W I T +NG D+ E K
Sbjct 151 ANEQETDRLTVNTELSQRALREIYLKPFEMVVKSANPWAIMTSYNKINGTHADSHEPLLK 210
Query 71 HLAKKEWD 78
+ + +W+
Sbjct 211 GILRGQWN 218
>gi|547243340|ref|WP_021979162.1| beta-N-acetylglucosaminidase [Clostridium sp. CAG:793]
gi|524123935|emb|CCZ00006.1| beta-N-acetylglucosaminidase [Clostridium sp. CAG:793]
Length=709
Score = 34.3 bits (77), Expect = 9.6, Method: Composition-based stats.
Identities = 22/72 (31%), Positives = 36/72 (50%), Gaps = 11/72 (15%)
Query 27 IEEREVSENGPFVLIRNKKNKWVITTCGALVNGKEFD------TKEDAEK-HLAKKEWDD 79
I E +S N P LI+N+ W+ TCG GK++D EDA K ++ + W +
Sbjct 86 IGESGISGNSPLSLIQNRTGAWICATCG----GKKYDNGTWSHASEDAIKYYMDPRNWLE 141
Query 80 ILTAALIFFTHV 91
++A+ F +
Sbjct 142 DDSSAIFQFLQI 153
>gi|591405264|gb|EXL40401.1| beta-glucosidase [Fusarium oxysporum f. sp. radicis-lycopersici
26381]
Length=842
Score = 34.3 bits (77), Expect = 9.8, Method: Composition-based stats.
Identities = 18/68 (26%), Positives = 33/68 (49%), Gaps = 2/68 (3%)
Query 13 SGKEEEEIKDVNLEIEEREVSEN--GPFVLIRNKKNKWVITTCGALVNGKEFDTKEDAEK 70
+ ++E + VN E+ +R + E PF ++ N W I T +NG D+ E K
Sbjct 151 ANEQETDRLTVNTELSQRALREIYLKPFEMVVKSANPWAIMTSYNKINGTHADSHEPLLK 210
Query 71 HLAKKEWD 78
+ + +W+
Sbjct 211 GILRGQWN 218
>gi|587724321|gb|EWZ95658.1| beta-glucosidase [Fusarium oxysporum f. sp. lycopersici MN25]
Length=841
Score = 34.3 bits (77), Expect = 9.9, Method: Composition-based stats.
Identities = 18/68 (26%), Positives = 33/68 (49%), Gaps = 2/68 (3%)
Query 13 SGKEEEEIKDVNLEIEEREVSEN--GPFVLIRNKKNKWVITTCGALVNGKEFDTKEDAEK 70
+ ++E + VN E+ +R + E PF ++ N W I T +NG D+ E K
Sbjct 151 ANEQETDRLTVNTELSQRALREIYLKPFEMVVKSANPWAIMTSYNKINGTHADSHEPLLK 210
Query 71 HLAKKEWD 78
+ + +W+
Sbjct 211 GILRGQWN 218
Lambda K H a alpha
0.311 0.130 0.361 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 438108004959