bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: All non-redundant GenBank CDS translations+PDB+SwissProt+PIR+PRF
excluding environmental samples from WGS projects
49,011,213 sequences; 17,563,301,199 total letters
Query= Contig-24_CDS_annotation_glimmer3.pl_2_3
Length=591
Score E
Sequences producing significant alignments: (Bits) Value
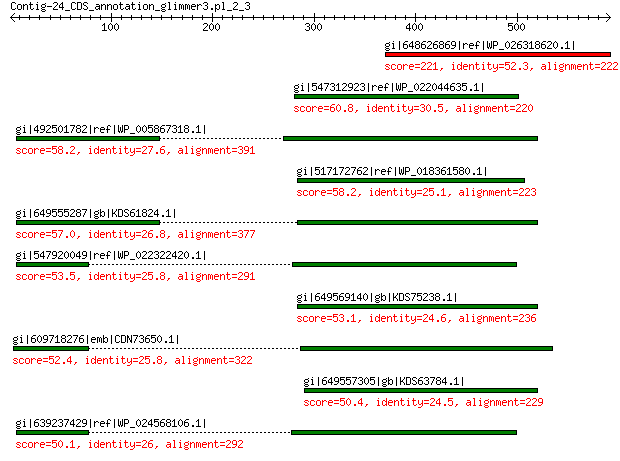
gi|648626869|ref|WP_026318620.1| hypothetical protein 221 6e-64
gi|547312923|ref|WP_022044635.1| putative uncharacterized protein 60.8 1e-06
gi|492501782|ref|WP_005867318.1| hypothetical protein 58.2 1e-05
gi|517172762|ref|WP_018361580.1| hypothetical protein 58.2 1e-05
gi|649555287|gb|KDS61824.1| capsid family protein 57.0 3e-05
gi|547920049|ref|WP_022322420.1| capsid protein VP1 53.5 3e-04
gi|649569140|gb|KDS75238.1| capsid family protein 53.1 4e-04
gi|609718276|emb|CDN73650.1| conserved hypothetical protein 52.4 8e-04
gi|649557305|gb|KDS63784.1| capsid family protein 50.4 0.001
gi|639237429|ref|WP_024568106.1| hypothetical protein 50.1 0.004
>gi|648626869|ref|WP_026318620.1| hypothetical protein [Alistipes onderdonkii]
Length=231
Score = 221 bits (562), Expect = 6e-64, Method: Compositional matrix adjust.
Identities = 116/232 (50%), Positives = 141/232 (61%), Gaps = 11/232 (5%)
Query 370 LSGGTRFRRRNYHFNDDGYFMEITSIVPRTYYPSYINPTSRQISLGQQYAPALDNIAMQG 429
+SGG F+RR +HFN+ GYFMEITS+VP YP+Y+NPT Q +LGQ+YAPALDNI MQ
Sbjct 1 MSGGDSFKRRTFHFNESGYFMEITSVVPTVMYPNYLNPTLLQTNLGQRYAPALDNIQMQP 60
Query 430 LKASTVFGEVQSLGATSPTYSNSV----------FAVPGFKLQDTNYVGYEPAWSELMTA 479
L T+ G S +YS+ + AV + VGY+PAW+ELMT
Sbjct 61 LTVPTLLGNAY-FNTGSGSYSHVLNHMGTGELRTVAVDKLSAAEGIAVGYQPAWAELMTG 119
Query 480 VSKPHGRLCNDLDYWVLSRDYGRNLASVMDTPAYKSFVSAAGAHVEELALQRLTAFFKRI 539
VSKPHGRLCNDLDYW R YG L S D F+ G V+ L ++ A+ K
Sbjct 120 VSKPHGRLCNDLDYWAFQRRYGTVLYSSNDAQDASVFLEELGNEVDTLDVETFNAWLKNT 179
Query 540 YVSPSSCPYILCGDFNYVFYDQRATAENFVLDNVADIVVFREKSKVNVATTL 591
YVS PYIL +NYVF D A+NFVLDN A+I V+REKSKVNV TL
Sbjct 180 YVSTDFVPYILPAMYNYVFADTDPNAQNFVLDNSAEISVYREKSKVNVPNTL 231
>gi|547312923|ref|WP_022044635.1| putative uncharacterized protein [Alistipes finegoldii CAG:68]
gi|524208404|emb|CCZ76639.1| putative uncharacterized protein [Alistipes finegoldii CAG:68]
Length=338
Score = 60.8 bits (146), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 67/243 (28%), Positives = 105/243 (43%), Gaps = 33/243 (14%)
Query 281 AVDVSTS-GNSVSMRNITFASRMQRYMDLAFAGGGRNSDFYESQFDVK-----LNQDNTC 334
A+D++ S G SV++ + +++Q +MD F GGR D + + + K +N+
Sbjct 41 ALDLNISTGFSVAVPELRLRTKIQNWMDRLFVSGGRVGDVFRTLWGTKSSAIYVNK---- 96
Query 335 PAFLGSDSFDMNANTLYQTTGFEDSSSPLGAFSGQLSGG-------TRFRRRNYHFNDDG 387
P FLG +N + + S+S A GQL+ + +Y+ + G
Sbjct 97 PDFLGVWQASINPSNVRAMA--NGSASGEDANLGQLAACVDRYCDFSGHSGIDYYAKEPG 154
Query 388 YFMEITSIVPRTYYPSYINPTSRQISLGQQYAPALDNIAMQGLKASTVFGEVQSLGAT-- 445
FM IT +VP Y ++P IS G + P L+ I Q + + T
Sbjct 155 TFMLITMLVPEPAYSQGLHPDLASISFGDDFNPELNGIGFQLVPRHRFSMMPRGFNFTGL 214
Query 446 ----SPTYSNSVFAVPGFKLQDTNY--VGYEPAWSELMTAVSKPHGRLCN--DLDYWVLS 497
SP + ++ V L D N VG E AWS L T S+ HG + YWVL+
Sbjct 215 DQEASPWFGHTGTGV----LVDPNMVSVGEEVAWSWLRTDYSRLHGDFAQNGNYQYWVLT 270
Query 498 RDY 500
R +
Sbjct 271 RRF 273
>gi|492501782|ref|WP_005867318.1| hypothetical protein [Parabacteroides distasonis]
gi|409230408|gb|EKN23272.1| hypothetical protein HMPREF1059_03257 [Parabacteroides distasonis
CL09T03C24]
Length=538
Score = 58.2 bits (139), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 62/256 (24%), Positives = 111/256 (43%), Gaps = 32/256 (13%)
Query 270 AWLKTSSFTDAAVDVSTSGNSVSMRNITFASRMQRYMDLAFAGGGRNSDFYESQFDVKLN 329
A+L+ +F V+V G VS+ ++ ++ +QR+ + G R + S F V+ +
Sbjct 280 AYLEPDNFQ---VNVDELG--VSINDLRTSNALQRWFERNARSGSRYIEQILSHFGVRSS 334
Query 330 QDN-TCPAFLGSDSFDMNANTLYQTTGFEDSSSPLGAFSGQ-LSGGTRFRRRNYHFNDDG 387
P FLG ++ + + QT+ DS+SP +G +S G + Y F + G
Sbjct 335 DARLQRPQFLGGGRTPISVSEVLQTSA-TDSTSPQANMAGHGISAGVNHGFKRY-FEEHG 392
Query 388 YFMEITSIVPRTYYPSYINPTSRQISLGQQYAPALDNIAMQGLKASTVFGEVQSLGATSP 447
Y + I SI PRT Y + R+ Y P ++ Q +K V+ + +P
Sbjct 393 YIIGIMSIRPRTGYQQGVPKDFRKFDNMDFYFPEFAHLGEQEIKNEEVYLQ------QTP 446
Query 448 TYSNSVFAVPGFKLQDTNYVGYEPAWSELMTAVSKPHGRLCNDLDYWVLSRDYGR----N 503
+N F GY P ++E ++++ HG ++ +W L+R + N
Sbjct 447 ASNNGTF-------------GYTPRYAEYKYSMNEVHGDFRGNMAFWHLNRIFSESPNLN 493
Query 504 LASVMDTPAYKSFVSA 519
V P+ + F +A
Sbjct 494 TTFVECNPSNRVFATA 509
Score = 54.7 bits (130), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 46/145 (32%), Positives = 64/145 (44%), Gaps = 28/145 (19%)
Query 7 KRNKKSRFKLFSGNPTSASWGTLIPTNVTRVVAGDDFSFQPGVGVQALPIVAPFMGSVCV 66
KR +++ F L N +A+ G L+P VV GD F + V+ P+VAP M V V
Sbjct 11 KRPRRNVFNLSYENKLTANAGELVPIMCKPVVPGDKFRVNTEMLVRLAPLVAPMMHRVDV 70
Query 67 KKEYFFIPDR-IYNVDRQLNFQGV--TDTPNTVYKPSIAPPIPFDISKPSGDAISFPVSA 123
YFF+P+R ++N +GV TDTP FP A
Sbjct 71 FTHYFFVPNRLLWNQWEDFITKGVDGTDTP------------------------VFPKIA 106
Query 124 LQTS-VPSGSLGAIVGPGSLADYMG 147
L+ V S ++ GSL DY+G
Sbjct 107 LRPDWVNPTSAAVLLDDGSLWDYLG 131
>gi|517172762|ref|WP_018361580.1| hypothetical protein [Prevotella nanceiensis]
Length=568
Score = 58.2 bits (139), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 56/238 (24%), Positives = 104/238 (44%), Gaps = 33/238 (14%)
Query 284 VSTSGNSVSMRNITFASRMQRYMDLAFAGGGRNSDFYESQFDVKLNQ--DNTCPAFLGSD 341
V + +S+ +I A +++ + G + E+ F + + + D C G D
Sbjct 297 VDSKRTMISVADIRNAFALEKLASVTMRAGKTYKEQMEAHFGISVEEGRDGRCTYIGGFD 356
Query 342 S----FDMNANTLYQTTGFEDSS------SPLGAFSGQLSGGTRFRRRNYHFNDDGYFME 391
S D+ ++ TG +D+S G +G SG RF + + G M
Sbjct 357 SNIQVGDVTQSSGTTVTGTKDTSFGGYLGRTTGKATGSGSGHIRFDAKEH-----GILMC 411
Query 392 ITSIVPRTYYPS-YINPTSRQISLGQQYAPALDNIAMQGLKASTVFGEVQSLGATSPTYS 450
I S+VP Y S ++P ++I G + P +N+ MQ L A + S Y+
Sbjct 412 IYSLVPDVQYDSKRVDPFVQKIERGDFFVPEFENLGMQPLFAKNI----------SYKYN 461
Query 451 NSVFAVPGFKLQDTNYVGYEPAWSELMTAVSKPHGRLCND--LDYWVLSRDYGRNLAS 506
N+ ++++ G++P +SE TA+ HG+ + L YW ++R G ++++
Sbjct 462 NNT---ANSRIKNLGAFGWQPRYSEYKTALDINHGQFVHQEPLSYWTVARARGESMSN 516
>gi|649555287|gb|KDS61824.1| capsid family protein [Parabacteroides distasonis str. 3999B
T(B) 4]
gi|649560568|gb|KDS66876.1| capsid family protein [Parabacteroides distasonis str. 3999B
T(B) 4]
gi|649561020|gb|KDS67307.1| capsid family protein [Parabacteroides distasonis str. 3999B
T(B) 4]
gi|649562724|gb|KDS68908.1| capsid family protein [Parabacteroides distasonis str. 3999B
T(B) 6]
Length=541
Score = 57.0 bits (136), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 43/142 (30%), Positives = 62/142 (44%), Gaps = 23/142 (16%)
Query 7 KRNKKSRFKLFSGNPTSASWGTLIPTNVTRVVAGDDFSFQPGVGVQALPIVAPFMGSVCV 66
KR +++ F L N + + G LIP VV GD F + V+ P+VAP M V V
Sbjct 11 KRPRRNVFNLSYENKLTVNAGELIPIMCKPVVPGDKFRVNTEMLVRLAPLVAPMMHRVDV 70
Query 67 KKEYFFIPDR-IYNVDRQLNFQGVTDTPNTVYKPSIAPPIPFDISKPSGDAISFPVSALQ 125
YFF+P+R I+N +GV T D+ FP +
Sbjct 71 FTHYFFVPNRLIWNKWEDFITKGVDGT----------------------DSPVFPTYSFP 108
Query 126 TSVPSGSLGAIVGPGSLADYMG 147
++V + + G GSL DY+G
Sbjct 109 STVDTANAHNSFGDGSLWDYLG 130
Score = 53.9 bits (128), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 58/242 (24%), Positives = 101/242 (42%), Gaps = 27/242 (11%)
Query 284 VSTSGNSVSMRNITFASRMQRYMDLAFAGGGRNSDFYESQFDVKLNQDN-TCPAFLGSDS 342
V+T V++ +I ++ +QR+ + G R + S F V+ + P FLG
Sbjct 292 VNTDQMGVNINDIRTSNALQRWFERNARSGSRYIEQILSHFGVRSSDARLQRPQFLGGGR 351
Query 343 FDMNANTLYQTTGFEDSSSPLGAFSGQ-LSGGTRFRRRNYHFNDDGYFMEITSIVPRTYY 401
++ + + QT+ DS+SP +G +S G Y F + GY M I SI PRT Y
Sbjct 352 TPISVSEVLQTSS-TDSTSPQANMAGHGISAGVNHGFTRY-FEEHGYIMGIMSIRPRTGY 409
Query 402 PSYINPTSRQISLGQQYAPALDNIAMQGLKASTVFGEVQSLGATSPTYSNSVFAVPGFKL 461
+ R+ Y P ++ Q +K ++ +S A T+
Sbjct 410 QQGVPKDFRKFDNMDFYFPEFAHLGEQEIKNEELYLN-ESDAANEGTF------------ 456
Query 462 QDTNYVGYEPAWSELMTAVSKPHGRLCNDLDYWVLSRDYGR----NLASVMDTPAYKSFV 517
GY P ++E + ++ HG ++ +W L+R + N V P+ + F
Sbjct 457 ------GYTPRYAEYKYSQNEVHGDFRGNMAFWHLNRIFKEKPNLNTTFVECNPSNRVFA 510
Query 518 SA 519
+A
Sbjct 511 TA 512
>gi|547920049|ref|WP_022322420.1| capsid protein VP1 [Parabacteroides merdae CAG:48]
gi|524592961|emb|CDD13573.1| capsid protein VP1 [Parabacteroides merdae CAG:48]
Length=553
Score = 53.5 bits (127), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 50/222 (23%), Positives = 94/222 (42%), Gaps = 23/222 (10%)
Query 279 DAAVDVSTSGNSVSMRNITFASRMQRYMDLAFAGGGRNSDFYESQFDVKLNQDN-TCPAF 337
+ + V+ +++ ++ ++ +QR+ + GG R + S F V+ + P F
Sbjct 299 NGTLKVNVDEMGININDLRTSNALQRWFERNARGGSRYIEQILSHFGVRSSDARLQRPQF 358
Query 338 LGSDSFDMNANTLYQTTGFEDSSSPLGAFSGQ-LSGGTRFRRRNYHFNDDGYFMEITSIV 396
LG ++ + + QT+ D +SP +G +S G ++Y F + GY + I SI
Sbjct 359 LGGGRMPISVSEVLQTSS-TDETSPQANMAGHGISAGINNGFKHY-FEEHGYIIGIMSIT 416
Query 397 PRTYYPSYINPTSRQISLGQQYAPALDNIAMQGLKASTVFGEVQSLGATSPTYSNSVFAV 456
PR+ Y + + Y P +++ Q +K +F + Y+N F
Sbjct 417 PRSGYQQGVPRDFTKFDNMDFYFPEFAHLSEQEIKNQELFV------SEDAAYNNGTF-- 468
Query 457 PGFKLQDTNYVGYEPAWSELMTAVSKPHGRLCNDLDYWVLSR 498
GY P ++E S+ HG +L +W L+R
Sbjct 469 -----------GYTPRYAEYKYHPSEAHGDFRGNLSFWHLNR 499
Score = 49.3 bits (116), Expect = 0.007, Method: Compositional matrix adjust.
Identities = 25/71 (35%), Positives = 39/71 (55%), Gaps = 0/71 (0%)
Query 7 KRNKKSRFKLFSGNPTSASWGTLIPTNVTRVVAGDDFSFQPGVGVQALPIVAPFMGSVCV 66
KR +++ F L + + + G L+P VV+GD F + V+ P+VAP M V V
Sbjct 11 KRPRRNAFNLSYESKLTLNMGELVPIMCMPVVSGDKFRVKTESLVRLAPLVAPMMHRVNV 70
Query 67 KKEYFFIPDRI 77
YFF+P+R+
Sbjct 71 FTHYFFVPNRL 81
>gi|649569140|gb|KDS75238.1| capsid family protein, partial [Parabacteroides distasonis str.
3999B T(B) 6]
Length=390
Score = 53.1 bits (126), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 58/242 (24%), Positives = 101/242 (42%), Gaps = 27/242 (11%)
Query 284 VSTSGNSVSMRNITFASRMQRYMDLAFAGGGRNSDFYESQFDVKLNQDN-TCPAFLGSDS 342
V+T V++ +I ++ +QR+ + G R + S F V+ + P FLG
Sbjct 141 VNTDQMGVNINDIRTSNALQRWFERNARSGSRYIEQILSHFGVRSSDARLQRPQFLGGGR 200
Query 343 FDMNANTLYQTTGFEDSSSPLGAFSGQ-LSGGTRFRRRNYHFNDDGYFMEITSIVPRTYY 401
++ + + QT+ DS+SP +G +S G Y F + GY M I SI PRT Y
Sbjct 201 TPISVSEVLQTSS-TDSTSPQANMAGHGISAGVNHGFTRY-FEEHGYIMGIMSIRPRTGY 258
Query 402 PSYINPTSRQISLGQQYAPALDNIAMQGLKASTVFGEVQSLGATSPTYSNSVFAVPGFKL 461
+ R+ Y P ++ Q +K ++ +S A T+
Sbjct 259 QQGVPKDFRKFDNMDFYFPEFAHLGEQEIKNEELYLN-ESDAANEGTF------------ 305
Query 462 QDTNYVGYEPAWSELMTAVSKPHGRLCNDLDYWVLSRDYGR----NLASVMDTPAYKSFV 517
GY P ++E + ++ HG ++ +W L+R + N V P+ + F
Sbjct 306 ------GYTPRYAEYKYSQNEVHGDFRGNMAFWHLNRIFKEKPNLNTTFVECNPSNRVFA 359
Query 518 SA 519
+A
Sbjct 360 TA 361
>gi|609718276|emb|CDN73650.1| conserved hypothetical protein [Elizabethkingia anophelis]
Length=537
Score = 52.4 bits (124), Expect = 8e-04, Method: Compositional matrix adjust.
Identities = 58/252 (23%), Positives = 105/252 (42%), Gaps = 35/252 (14%)
Query 287 SGNSVSMRNITFASRMQRYMDLAFAGGGRNSDFYESQFDVKLNQDNTC-PAFLGSDSFDM 345
S N ++ ++ A ++Q +++ G R ++ S F VK + P FLG + +
Sbjct 285 SENVSTVNDLRRAFKLQEWLEKNARAGSRYAESILSFFGVKTSDGRLQRPEFLGGNKSPI 344
Query 346 NANTLYQTTGFEDSSSPLGAFSGQLSGGTRFRRRNYHFNDDGYFMEITSIVPRTYYPSYI 405
+ + Q + DS++P G +G G + + F + GY + + S++P+T SY
Sbjct 345 MISEVLQQSA-TDSTTPQGNMAGHGIGIGKDGGFSRFFEEHGYVIGLMSVIPKT---SYS 400
Query 406 NPTSRQISLGQQYA---PALDNIAMQGLKASTVFGEVQSLGATSPTYSNSVFAVPGFKLQ 462
R S ++ P ++I Q P Y+ +FA
Sbjct 401 QGIPRHFSKSDKFDYFWPQFEHIGEQ------------------PVYNKEIFAKNIDAFD 442
Query 463 DTNYVGYEPAWSELMTAVSKPHGRLCNDLDYWVLSRDYGRNLASVMDTPAYKSFVSAAGA 522
GY P +SE + S HG +DL +W L R + + V++ +SF+
Sbjct 443 SEAVFGYLPRYSEYKFSPSTVHGDFKDDLYFWHLGRIFDTDKPPVLN----QSFIEC--- 495
Query 523 HVEELALQRLTA 534
++ AL R+ A
Sbjct 496 --DKNALSRIFA 505
Score = 49.3 bits (116), Expect = 0.007, Method: Compositional matrix adjust.
Identities = 25/74 (34%), Positives = 38/74 (51%), Gaps = 0/74 (0%)
Query 4 LSRKRNKKSRFKLFSGNPTSASWGTLIPTNVTRVVAGDDFSFQPGVGVQALPIVAPFMGS 63
++ K K S F + S ++G L+P + V+ GD S P + P++AP M
Sbjct 7 VASKAPKSSTFNMSYDRKFSMNFGDLVPIHCQEVIPGDKISINPQHMTRLAPMIAPVMHE 66
Query 64 VCVKKEYFFIPDRI 77
V V YFF+P+RI
Sbjct 67 VNVFIHYFFVPNRI 80
>gi|649557305|gb|KDS63784.1| capsid family protein [Parabacteroides distasonis str. 3999B
T(B) 4]
gi|649559156|gb|KDS65543.1| capsid family protein [Parabacteroides distasonis str. 3999B
T(B) 6]
Length=245
Score = 50.4 bits (119), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 56/235 (24%), Positives = 98/235 (42%), Gaps = 27/235 (11%)
Query 291 VSMRNITFASRMQRYMDLAFAGGGRNSDFYESQFDVKLNQDN-TCPAFLGSDSFDMNANT 349
V++ +I ++ +QR+ + G R + S F V+ + P FLG ++ +
Sbjct 3 VNINDIRTSNALQRWFERNARSGSRYIEQILSHFGVRSSDARLQRPQFLGGGRTPISVSE 62
Query 350 LYQTTGFEDSSSPLGAFSGQ-LSGGTRFRRRNYHFNDDGYFMEITSIVPRTYYPSYINPT 408
+ QT+ DS+SP +G +S G Y F + GY M I SI PRT Y +
Sbjct 63 VLQTSS-TDSTSPQANMAGHGISAGVNHGFTRY-FEEHGYIMGIMSIRPRTGYQQGVPKD 120
Query 409 SRQISLGQQYAPALDNIAMQGLKASTVFGEVQSLGATSPTYSNSVFAVPGFKLQDTNYVG 468
R+ Y P ++ Q +K ++ +S A T+ G
Sbjct 121 FRKFDNMDFYFPEFAHLGEQEIKNEELYLN-ESDAANEGTF------------------G 161
Query 469 YEPAWSELMTAVSKPHGRLCNDLDYWVLSRDYGR----NLASVMDTPAYKSFVSA 519
Y P ++E + ++ HG ++ +W L+R + N V P+ + F +A
Sbjct 162 YTPRYAEYKYSQNEVHGDFRGNMAFWHLNRIFKEKPNLNTTFVECNPSNRVFATA 216
>gi|639237429|ref|WP_024568106.1| hypothetical protein [Elizabethkingia anophelis]
Length=546
Score = 50.1 bits (118), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 51/223 (23%), Positives = 93/223 (42%), Gaps = 23/223 (10%)
Query 278 TDAAVDVSTSGNSVSMRNITFASRMQRYMDLAFAGGGRNSDFYESQFDVKLNQDNTC-PA 336
++ VD+ T+ S ++ ++ A ++Q +++ G R ++ S F VK + P
Sbjct 286 SNLGVDLKTASGS-TINDLRRAFKLQEWLEKNARAGSRYAESILSFFGVKTSDGRLQRPE 344
Query 337 FLGSDSFDMNANTLYQTTGFEDSSSPLGAFSGQ-LSGGTRFRRRNYHFNDDGYFMEITSI 395
FLG + + + + Q + DS++P G +G +S G + F + GY + + S+
Sbjct 345 FLGGNKTPILISEVLQQSS-TDSTTPQGNMAGHGISVGKEGGFSKF-FEEHGYVIGLMSV 402
Query 396 VPRTYYPSYINPTSRQISLGQQYAPALDNIAMQGLKASTVFGEVQSLGATSPTYSNSVFA 455
+P+T Y I + + P ++I Q P Y+ +FA
Sbjct 403 IPKTSYSQGIPRHFSKFDKFDYFWPQFEHIGEQ------------------PVYNKEIFA 444
Query 456 VPGFKLQDTNYVGYEPAWSELMTAVSKPHGRLCNDLDYWVLSR 498
GY P +SE + S HG + L +W L R
Sbjct 445 KNVGDYDSGGVFGYVPRYSEYKYSPSTIHGDFKDTLYFWHLGR 487
Score = 48.1 bits (113), Expect = 0.015, Method: Compositional matrix adjust.
Identities = 25/71 (35%), Positives = 36/71 (51%), Gaps = 0/71 (0%)
Query 7 KRNKKSRFKLFSGNPTSASWGTLIPTNVTRVVAGDDFSFQPGVGVQALPIVAPFMGSVCV 66
K K S F + S ++G L+P + +V GD S P + P++AP M V V
Sbjct 10 KAPKSSTFNMSYDRKFSMNFGDLVPIHCQEIVPGDKISINPQHMTRLAPMLAPVMHEVNV 69
Query 67 KKEYFFIPDRI 77
YFF+P+RI
Sbjct 70 FIHYFFVPNRI 80
Lambda K H a alpha
0.318 0.134 0.396 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 4376806011489