bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: All non-redundant GenBank CDS translations+PDB+SwissProt+PIR+PRF
excluding environmental samples from WGS projects
49,011,213 sequences; 17,563,301,199 total letters
Query= Contig-25_CDS_annotation_glimmer3.pl_2_6
Length=76
Score E
Sequences producing significant alignments: (Bits) Value
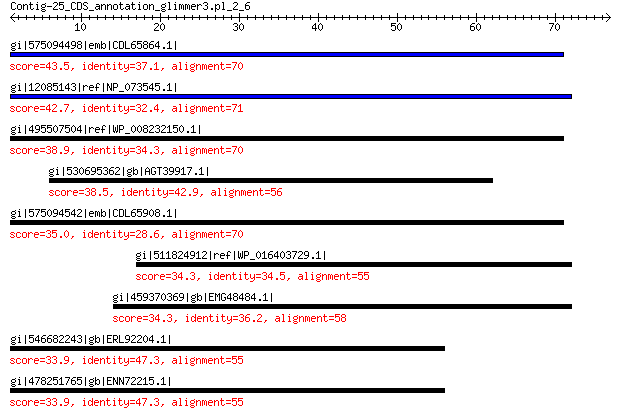
gi|575094498|emb|CDL65864.1| unnamed protein product 43.5 8e-04
gi|12085143|ref|NP_073545.1| nonstructural protein 42.7 0.001
gi|495507504|ref|WP_008232150.1| hypothetical protein 38.9 0.027
gi|530695362|gb|AGT39917.1| packaging protein 38.5 0.040
gi|575094542|emb|CDL65908.1| unnamed protein product 35.0 0.53
gi|511824912|ref|WP_016403729.1| glucose-1-phosphate thymidylylt... 34.3 4.2
gi|459370369|gb|EMG48484.1| putative transporter 34.3 6.1
gi|546682243|gb|ERL92204.1| hypothetical protein D910_09524 33.9 9.2
gi|478251765|gb|ENN72215.1| hypothetical protein YQE_11145 33.9 9.3
>gi|575094498|emb|CDL65864.1| unnamed protein product [uncultured bacterium]
Length=92
Score = 43.5 bits (101), Expect = 8e-04, Method: Compositional matrix adjust.
Identities = 26/76 (34%), Positives = 47/76 (62%), Gaps = 7/76 (9%)
Query 1 MVYKIYSVKDAVAGQFSEPRIFMNEGLAVRWFKNLCE--KSEIAS---DLSLYYLGEYDL 55
M+ + YS+KD G F P N+ LA+R F+++ + S I+S D SL+ +G +D
Sbjct 1 MILQAYSIKDDKVG-FYAPTYEQNDKLAIRVFQSIYKDKNSTISSYPADFSLFAVGSFDT 59
Query 56 ESGVISS-APEFVMNG 70
++GV++S P+F+++
Sbjct 60 DTGVLTSQTPKFLVSA 75
>gi|12085143|ref|NP_073545.1| nonstructural protein [Bdellovibrio phage phiMH2K]
gi|75089166|sp|Q9G052.1|C_BPPHM RecName: Full=Protein VP5 [Bdellovibrio phage phiMH2K]
gi|12017991|gb|AAG45347.1|AF306496_8 Vp5 [Bdellovibrio phage phiMH2K]
Length=84
Score = 42.7 bits (99), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 23/78 (29%), Positives = 42/78 (54%), Gaps = 7/78 (9%)
Query 1 MVYKIYSVKDAVAGQFSEPRIFMNEGLAVRWFKNLCE--KSEIAS---DLSLYYLGEYDL 55
M K++S++D+ G + P G A R F+ L + +S +A+ D L++LGEYD
Sbjct 1 MQLKVFSIRDSKTGVYGTPFYQHTHGQAERSFQQLAKDPQSTVANHPEDFDLFHLGEYDD 60
Query 56 ESGVIS--SAPEFVMNGV 71
++G ++ PE + +
Sbjct 61 QTGKLTPLDTPEHCVKAI 78
>gi|495507504|ref|WP_008232150.1| hypothetical protein [Richelia intracellularis]
gi|471331138|emb|CCH66546.1| hypothetical protein RINTHH_3910 [Richelia intracellularis HH01]
Length=86
Score = 38.9 bits (89), Expect = 0.027, Method: Compositional matrix adjust.
Identities = 24/79 (30%), Positives = 37/79 (47%), Gaps = 9/79 (11%)
Query 1 MVYKIYSVKDAVAGQFSEPRIFMNEGLAVRWFKNLCEKSEIAS-------DLSLYYLGEY 53
M + V+D A QF P ++G+A+R F+++ + S D LY LGE+
Sbjct 1 MSKPVVCVRDVKADQFGNPXFPSSKGVALREFQDVVNQPNEDSIIYKHPEDFDLYLLGEF 60
Query 54 DLESGVIS--SAPEFVMNG 70
D +G PE +M G
Sbjct 61 DESTGKFDVLDLPELMMAG 79
>gi|530695362|gb|AGT39917.1| packaging protein [Marine gokushovirus]
Length=83
Score = 38.5 bits (88), Expect = 0.040, Method: Compositional matrix adjust.
Identities = 24/62 (39%), Positives = 34/62 (55%), Gaps = 6/62 (10%)
Query 6 YSVKDAVAGQFSEPRIFMNEGLAVRWFKNLCEKSE------IASDLSLYYLGEYDLESGV 59
Y+V D A FS P + + +G A+R ++L S SD SL+ LGE+D SGV
Sbjct 6 YAVYDRKAELFSAPFLEIKDGTAIRAIQDLVVNSPDHAFATHPSDFSLHKLGEFDDVSGV 65
Query 60 IS 61
I+
Sbjct 66 IT 67
>gi|575094542|emb|CDL65908.1| unnamed protein product [uncultured bacterium]
Length=80
Score = 35.0 bits (79), Expect = 0.53, Method: Compositional matrix adjust.
Identities = 20/76 (26%), Positives = 38/76 (50%), Gaps = 7/76 (9%)
Query 1 MVYKIYSVKDAVAGQFSEPRIFMNEGLAVRWFKNLCEKSEIA-----SDLSLYYLGEYDL 55
M+Y +Y ++D V + P ++ A R+F++ + S D +LY +G YD
Sbjct 1 MIYNLYVIRD-VKSCYGVPLAMESDAFAARYFEHEIQVSGTVMSTHYRDFALYCIGTYDA 59
Query 56 E-SGVISSAPEFVMNG 70
+ + +I+ P V+ G
Sbjct 60 DKATIITDTPRLVIEG 75
>gi|511824912|ref|WP_016403729.1| glucose-1-phosphate thymidylyltransferase [Agarivorans albus]
gi|511670110|dbj|GAD03962.1| glucose-1-phosphate thymidylyltransferase [Agarivorans albus
MKT 106]
Length=226
Score = 34.3 bits (77), Expect = 4.2, Method: Compositional matrix adjust.
Identities = 19/64 (30%), Positives = 33/64 (52%), Gaps = 9/64 (14%)
Query 17 SEPRIFMNEGLAVRW-FKNLCEKSEIASDLSLYYL--------GEYDLESGVISSAPEFV 67
SE + +N + W ++ +C+ S+ SD LY + G++ LE G++ PEF
Sbjct 102 SEQFLVINGDVWNDWGYQAICQNSQTDSDAHLYLVTNPSHNLTGDFSLEQGLVIDKPEFT 161
Query 68 MNGV 71
+GV
Sbjct 162 FSGV 165
>gi|459370369|gb|EMG48484.1| putative transporter [Candida maltosa Xu316]
Length=575
Score = 34.3 bits (77), Expect = 6.1, Method: Compositional matrix adjust.
Identities = 21/60 (35%), Positives = 28/60 (47%), Gaps = 4/60 (7%)
Query 14 GQFSEPRIFMNEGLAVRWFKNLCEKSEIASDLSLYYLGEY--DLESGVISSAPEFVMNGV 71
G F P L W+K E+A + YYLG+Y L SG++S A E M G+
Sbjct 209 GAFEAPSYLAYHALFASWYK--ASTGEVARRAAFYYLGQYLGILTSGLLSGAIERHMGGI 266
>gi|546682243|gb|ERL92204.1| hypothetical protein D910_09524 [Dendroctonus ponderosae]
Length=1125
Score = 33.9 bits (76), Expect = 9.2, Method: Composition-based stats.
Identities = 26/70 (37%), Positives = 33/70 (47%), Gaps = 15/70 (21%)
Query 1 MVYKIYSVKDAVAGQFSEPRIFMNEGLA------VRWFKNLCEKS---------EIASDL 45
M+ +IY++K A A S+P ++ L VR F NL KS EI L
Sbjct 26 MLTRIYNIKKACADSKSKPAFLSDKNLESSIKSIVRRFPNLDAKSLIPIQGLRNEIIKSL 85
Query 46 SLYYLGEYDL 55
SLYYL DL
Sbjct 86 SLYYLTFVDL 95
>gi|478251765|gb|ENN72215.1| hypothetical protein YQE_11145, partial [Dendroctonus ponderosae]
Length=1125
Score = 33.9 bits (76), Expect = 9.3, Method: Composition-based stats.
Identities = 26/70 (37%), Positives = 33/70 (47%), Gaps = 15/70 (21%)
Query 1 MVYKIYSVKDAVAGQFSEPRIFMNEGLA------VRWFKNLCEKS---------EIASDL 45
M+ +IY++K A A S+P ++ L VR F NL KS EI L
Sbjct 26 MLTRIYNIKKACADSKSKPAFLSDKNLESSIKSIVRRFPNLDAKSLIPIQGLRNEIIKSL 85
Query 46 SLYYLGEYDL 55
SLYYL DL
Sbjct 86 SLYYLTFVDL 95
Lambda K H a alpha
0.318 0.135 0.386 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 442533451452