bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: All non-redundant GenBank CDS translations+PDB+SwissProt+PIR+PRF
excluding environmental samples from WGS projects
49,011,213 sequences; 17,563,301,199 total letters
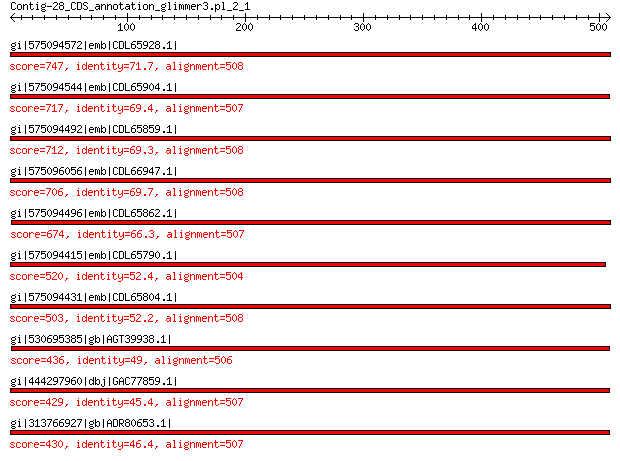
Query= Contig-28_CDS_annotation_glimmer3.pl_2_1
Length=508
Score E
Sequences producing significant alignments: (Bits) Value
gi|575094572|emb|CDL65928.1| unnamed protein product 747 0.0
gi|575094544|emb|CDL65904.1| unnamed protein product 717 0.0
gi|575094492|emb|CDL65859.1| unnamed protein product 712 0.0
gi|575096056|emb|CDL66947.1| unnamed protein product 706 0.0
gi|575094496|emb|CDL65862.1| unnamed protein product 674 0.0
gi|575094415|emb|CDL65790.1| unnamed protein product 520 3e-176
gi|575094431|emb|CDL65804.1| unnamed protein product 503 8e-170
gi|530695385|gb|AGT39938.1| major capsid protein 436 4e-144
gi|444297960|dbj|GAC77859.1| major capsid protein 429 1e-141
gi|313766927|gb|ADR80653.1| putative major coat protein 430 1e-141
>gi|575094572|emb|CDL65928.1| unnamed protein product [uncultured bacterium]
Length=556
Score = 747 bits (1929), Expect = 0.0, Method: Compositional matrix adjust.
Identities = 364/516 (71%), Positives = 416/516 (81%), Gaps = 10/516 (2%)
Query 1 LDEVLPGDTFKIKTSKVVRLQTLITPMMDNLYLDTYFFFVPNRLVWSHWKEFNGENTQSA 60
++EVLPGDTFK+KTSKV+RLQTL+TPMMDN+YLDTY+FFVPNRLVW HWKEFNGENTQSA
Sbjct 43 IEEVLPGDTFKVKTSKVIRLQTLLTPMMDNIYLDTYYFFVPNRLVWEHWKEFNGENTQSA 102
Query 61 WLPTTEYEIPQITAPATGGWSIGTIADYLGIPTGVPDLSVNALPFRAYALVMNEWFRDEN 120
W+P EY+IPQ+TAP GGW+IGT+ADY GIPTGV +SVNALPFRAYALV NEWFRD+N
Sbjct 103 WIPEVEYQIPQLTAPE-GGWNIGTLADYFGIPTGVSGISVNALPFRAYALVCNEWFRDQN 161
Query 121 LTDPLVVPLDDATVAGVNTGAYVTDVAKGGLPFVAAKYHDYFTSCLPAPQKGPDVVIPGG 180
L+DPL +P+ DATV GVNTG ++TDV KGGLP+ AAKYHDYFTSCLPAPQKGPDV IP
Sbjct 162 LSDPLNIPVGDATVTGVNTGTFITDVVKGGLPYTAAKYHDYFTSCLPAPQKGPDVTIPVT 221
Query 181 TGMSVPVI---PQADKVPSGLITMPYTATFLNETPVRSTTGIFFNDSGSQTNGVSAGSSE 237
+G ++PV+ D P + + L + + ++ V GS
Sbjct 222 SGHNLPVMFLNETHDAGPYKPFGVGIQNSELRNFYGFGSGSSGATSTSDTSSTVEVGSDG 281
Query 238 DALP----VIDNLWAVGDG-VATATINQLRLAFQIQKLYEKDARGGTRYTEILRSHFGVT 292
+ N+WAV G V ATINQLRLAFQ+QKLYEKDARGGTRYTEI+RSHFGV
Sbjct 282 TGIGQNFWTPTNMWAVESGDVGMATINQLRLAFQLQKLYEKDARGGTRYTEIIRSHFGVV 341
Query 293 SPDSRLQRPEYLGGNRIPIRINQIVQQSATQEGSTPQGNPVGLSLTSDNHGDFTKSFTEH 352
SPDSRLQRPEYLGGNRIPI +NQI+QQS + E S P G G+S+T+D + DF KSF EH
Sbjct 342 SPDSRLQRPEYLGGNRIPINVNQIIQQSQSTEQS-PLGALAGMSVTTDKNSDFIKSFVEH 400
Query 353 GFILGLMVARYDHTYQQGLDRMFSRKSRFDYYWPVFANIGEQAVLNKEIYAQGTNEDDEV 412
G+I+GL+VARYDHTYQQGLDRM+SRK RFD+YWPV ANIGEQAVLNKEIY G++ DDEV
Sbjct 401 GYIIGLVVARYDHTYQQGLDRMWSRKDRFDFYWPVLANIGEQAVLNKEIYIDGSDTDDEV 460
Query 413 FGYQEAWADYRYKPNRVCGEMRSQAPQSLDVWHLGDDYSKLPSLSDEWIREDKTNVDRVL 472
FGYQEAWA+YRYKPNRVCGEMRS APQSLDVWHLGDDYS LP LSD WIREDKTNVDRVL
Sbjct 461 FGYQEAWAEYRYKPNRVCGEMRSSAPQSLDVWHLGDDYSSLPYLSDSWIREDKTNVDRVL 520
Query 473 AVQSSVSNQLFADIYVQNRCTRPMPMYSIPGLIDHH 508
AV SSVS+QLFADIY+ N+ TRPMPMYSIPGLIDHH
Sbjct 521 AVTSSVSDQLFADIYICNKATRPMPMYSIPGLIDHH 556
>gi|575094544|emb|CDL65904.1| unnamed protein product [uncultured bacterium]
Length=551
Score = 717 bits (1850), Expect = 0.0, Method: Compositional matrix adjust.
Identities = 352/516 (68%), Positives = 406/516 (79%), Gaps = 16/516 (3%)
Query 1 LDEVLPGDTFKIKTSKVVRLQTLITPMMDNLYLDTYFFFVPNRLVWSHWKEFNGENTQSA 60
+DEVLPGDTF +K+SKV+R+Q+L+TP+MDN+YLDTY+FFVPNRLVWSHW++FNGENT+SA
Sbjct 42 IDEVLPGDTFNVKSSKVIRMQSLVTPIMDNIYLDTYYFFVPNRLVWSHWQQFNGENTESA 101
Query 61 WLPTTEYEIPQITAPATGGWSIGTIADYLGIPTGVPDLSVNALPFRAYALVMNEWFRDEN 120
WLPTTEY++PQ+TAPA GWSIGTIADY GIPTGV SVNALPFRAYAL+ NEWFRDEN
Sbjct 102 WLPTTEYQVPQVTAPA-NGWSIGTIADYFGIPTGVA-CSVNALPFRAYALICNEWFRDEN 159
Query 121 LTDPLVVPLDDATVAGVNTGAYVTDVAKGGLPFVAAKYHDYFTSCLPAPQKGPDVVIPGG 180
L+DPL +P+ DATV G N Y+TD+ KGG+PF A KYHDYFTSCLPAPQKGPDV++P
Sbjct 160 LSDPLNIPISDATVVGSNGDNYITDIVKGGMPFKACKYHDYFTSCLPAPQKGPDVLLPLS 219
Query 181 TGMSVPVIPQADKV-PSGLITMPYTAT---FLNETPVRSTTGIFFNDSGSQTNGVSAGSS 236
+ VPV V P P L+ T +R+ F G +
Sbjct 220 SS-PVPVTTSDTMVDPLQYSKYPMAGVDSWNLSPTLMRNIIRPF---EGVEGANYQVHQF 275
Query 237 EDALPVID-----NLWAVGDGVATATINQLRLAFQIQKLYEKDARGGTRYTEILRSHFGV 291
+P ID NL A A+INQLRLAFQIQ+LYE+DARGGTRY EIL+SHFGV
Sbjct 276 TGDIPTIDAFRPLNLVANLQNATAASINQLRLAFQIQRLYERDARGGTRYIEILKSHFGV 335
Query 292 TSPDSRLQRPEYLGGNRIPIRINQIVQQSATQEGSTPQGNPVGLSLTSDNHGDFTKSFTE 351
TSPD+RLQRPEYLGGNRIPI INQ++QQS T ++PQGNPVG SLT+D + DF KSF E
Sbjct 336 TSPDARLQRPEYLGGNRIPININQVLQQSET-TSTSPQGNPVGQSLTTDTNADFVKSFVE 394
Query 352 HGFILGLMVARYDHTYQQGLDRMFSRKSRFDYYWPVFANIGEQAVLNKEIYAQGTNEDDE 411
HGF++GLMVARYDHTYQQGL+R +SRK RFDYYWPVFA+IGEQAVLNKEIY GT DDE
Sbjct 395 HGFVIGLMVARYDHTYQQGLERFWSRKDRFDYYWPVFAHIGEQAVLNKEIYTSGTAVDDE 454
Query 412 VFGYQEAWADYRYKPNRVCGEMRSQAPQSLDVWHLGDDYSKLPSLSDEWIREDKTNVDRV 471
VFGYQEA+ADYRYKP+RV GEMRS APQSLDVWHL DDY+ LPSLSD WIRE + VDRV
Sbjct 455 VFGYQEAYADYRYKPSRVTGEMRSAAPQSLDVWHLADDYASLPSLSDSWIRESASTVDRV 514
Query 472 LAVQSSVSNQLFADIYVQNRCTRPMPMYSIPGLIDH 507
LAV S+VS QLF DIY+QNR TRPMPMYS+PGLIDH
Sbjct 515 LAVSSNVSAQLFCDIYIQNRSTRPMPMYSVPGLIDH 550
>gi|575094492|emb|CDL65859.1| unnamed protein product [uncultured bacterium]
Length=551
Score = 712 bits (1838), Expect = 0.0, Method: Compositional matrix adjust.
Identities = 352/518 (68%), Positives = 405/518 (78%), Gaps = 19/518 (4%)
Query 1 LDEVLPGDTFKIKTSKVVRLQTLITPMMDNLYLDTYFFFVPNRLVWSHWKEFNGENTQSA 60
+DE+LPGDTF I TSKVVR+Q+L+TP+MDN+YLDTYFFFVPNRL WSHW+E GENTQSA
Sbjct 43 VDEILPGDTFSIDTSKVVRMQSLLTPVMDNIYLDTYFFFVPNRLTWSHWRELMGENTQSA 102
Query 61 WLPTTEYEIPQITAPATGGWSIGTIADYLGIPTGVPDLSVNALPFRAYALVMNEWFRDEN 120
W P EY +PQITAP GGW++GTIADY+GIPTGV LSVNA+PFRAYAL+ NEWFRDEN
Sbjct 103 WTPQVEYSVPQITAPE-GGWNVGTIADYMGIPTGVSGLSVNAMPFRAYALICNEWFRDEN 161
Query 121 LTDPLVVPLDDATVAGVNTGAYVTDVAKGGLPFVAAKYHDYFTSCLPAPQKGPDVVIPG- 179
LTDPL +P+ DATVAGVNTG YVTDVAKGGLPF AAKYHDYFTSCLPAPQKGPDV+I
Sbjct 162 LTDPLNIPVGDATVAGVNTGTYVTDVAKGGLPFKAAKYHDYFTSCLPAPQKGPDVLISAV 221
Query 180 GTGMSVPVIPQADKVPSGLITMPYTATFLNETPVRSTTGIFFNDSGSQTNGVSAGSSEDA 239
G+G+ VPV + S + P N S+T + + G V + + +
Sbjct 222 GSGI-VPVTATDNDNDSLNVNSPGMRFVGN-----SSTSVNYLAFGGGDGYVVTDTPKPS 275
Query 240 LPVI------DNLWA---VGDGVATATINQLRLAFQIQKLYEKDARGGTRYTEILRSHFG 290
P+ NLWA + ATINQLR AFQIQKLYE+DARGGTRY EIL+SHFG
Sbjct 276 TPIHGISMIPTNLWADLSTATDLPVATINQLRTAFQIQKLYERDARGGTRYIEILKSHFG 335
Query 291 VTSPDSRLQRPEYLGGNRIPIRINQIVQQSATQEGSTPQGNPVGLSLTSDNHGDFTKSFT 350
VTSPD+RLQRPEYLGG+R+PI INQ++Q S T G+TPQGN SLT+D+H +FTKSF
Sbjct 336 VTSPDARLQRPEYLGGSRVPININQVIQSSET--GATPQGNAAAYSLTTDSHSEFTKSFV 393
Query 351 EHGFILGLMVARYDHTYQQGLDRMFSRKSRFDYYWPVFANIGEQAVLNKEIYAQGTNEDD 410
EHGFI+GLMVARYDH+YQQGL R +SRK RFDYYWPVFAN+GE AV NKEI+AQGT+ DD
Sbjct 394 EHGFIIGLMVARYDHSYQQGLQRFWSRKDRFDYYWPVFANLGEMAVKNKEIFAQGTDVDD 453
Query 411 EVFGYQEAWADYRYKPNRVCGEMRSQAPQSLDVWHLGDDYSKLPSLSDEWIREDKTNVDR 470
EVFGYQEAWADYRYKP+ V GEMRSQ QSLD+WHL DDY LPSLSD WIRED + V+R
Sbjct 454 EVFGYQEAWADYRYKPSVVTGEMRSQYAQSLDIWHLADDYENLPSLSDSWIREDSSTVNR 513
Query 471 VLAVQSSVSNQLFADIYVQNRCTRPMPMYSIPGLIDHH 508
VLAV SVS QLF DIY++ TRPMP+YSIPGLIDHH
Sbjct 514 VLAVSDSVSAQLFCDIYIRCLATRPMPLYSIPGLIDHH 551
>gi|575096056|emb|CDL66947.1| unnamed protein product [uncultured bacterium]
Length=570
Score = 706 bits (1823), Expect = 0.0, Method: Compositional matrix adjust.
Identities = 354/530 (67%), Positives = 407/530 (77%), Gaps = 24/530 (5%)
Query 1 LDEVLPGDTFKIKTSKVVRLQTLITPMMDNLYLDTYFFFVPNRLVWSHWKEFNGENTQSA 60
L+EVLPGDTF + +SKVVR+QTL+TPMMDN+YLDTY+FFVPNRLVW HWKEF GEN +SA
Sbjct 43 LEEVLPGDTFSVDSSKVVRMQTLLTPMMDNVYLDTYYFFVPNRLVWQHWKEFCGENNESA 102
Query 61 WLPTTEYEIPQITAPATGGWSIGTIADYLGIPTGVPDLSVNALPFRAYALVMNEWFRDEN 120
W+P TEY IPQ+ +P GG+ +GTIADY G+PTGV +LSV+ALPFRAYAL+MNEWFRDEN
Sbjct 103 WIPQTEYAIPQLKSPV-GGFEVGTIADYFGLPTGVANLSVSALPFRAYALIMNEWFRDEN 161
Query 121 LTDPLVVPLDDATVAGVNTGAYVTDVAKGGLPFVAAKYHDYFTSCLPAPQKGPDVVIP-- 178
L DPLVVP DDATV GVNTG +VTDVAKGG PFVAAKYHDYFTS LPAPQKGPDVVIP
Sbjct 162 LMDPLVVPTDDATVTGVNTGIFVTDVAKGGKPFVAAKYHDYFTSALPAPQKGPDVVIPVA 221
Query 179 ---------GGTGMSVPVIPQADKVPSGLITMPYTATFLNETPVRSTTGIFFNDSGS--- 226
G G+++ + + +GL T L + + + GS
Sbjct 222 SAGNYNVVGNGKGLALSDGSKMSIICNGLSGSNGQGTELFASGILGSQVGSSGGFGSGSS 281
Query 227 -QTNGVSAGSSEDALPVIDNLWAVG------DGVATATINQLRLAFQIQKLYEKDARGGT 279
+ +G+ G A + +NL G A ATINQLR+AFQIQK YEK ARGG+
Sbjct 282 LRGDGIILGVPT-AAQLGNNLENSGLIAIASGNAAAATINQLRMAFQIQKFYEKQARGGS 340
Query 280 RYTEILRSHFGVTSPDSRLQRPEYLGGNRIPIRINQIVQQSATQEGST-PQGNPVGLSLT 338
RYTE++RS FGVTSPD+RLQR EYLGGNRIPI INQ++QQS T ST PQG VG+S T
Sbjct 341 RYTEVIRSFFGVTSPDARLQRSEYLGGNRIPININQVIQQSGTGSASTTPQGTVVGMSQT 400
Query 339 SDNHGDFTKSFTEHGFILGLMVARYDHTYQQGLDRMFSRKSRFDYYWPVFANIGEQAVLN 398
+D H DFTKSFTEHGFI+G+M ARYDHTYQQG+DRM+SRK +FDYYWPVF+NIGEQA+ N
Sbjct 401 TDTHSDFTKSFTEHGFIIGVMCARYDHTYQQGIDRMWSRKDKFDYYWPVFSNIGEQAIKN 460
Query 399 KEIYAQGTNEDDEVFGYQEAWADYRYKPNRVCGEMRSQAPQSLDVWHLGDDYSKLPSLSD 458
KEIYAQG DDEVFGYQEAWA+YRYKP+RV GEMRS QSLDVWHL DDYSKLPSLSD
Sbjct 461 KEIYAQGNATDDEVFGYQEAWAEYRYKPSRVTGEMRSSYAQSLDVWHLADDYSKLPSLSD 520
Query 459 EWIREDKTNVDRVLAVQSSVSNQLFADIYVQNRCTRPMPMYSIPGLIDHH 508
EWIRED ++RVLAV SNQ FADIYV+N CTRPMPMYSIPGLIDHH
Sbjct 521 EWIREDAKTLNRVLAVSDQNSNQFFADIYVKNLCTRPMPMYSIPGLIDHH 570
>gi|575094496|emb|CDL65862.1| unnamed protein product [uncultured bacterium]
Length=568
Score = 674 bits (1739), Expect = 0.0, Method: Compositional matrix adjust.
Identities = 336/533 (63%), Positives = 398/533 (75%), Gaps = 33/533 (6%)
Query 2 DEVLPGDTFKIKTSKVVRLQTLITPMMDNLYLDTYFFFVPNRLVWSHWKEFNGENTQSAW 61
DEVLPGDTF++KT+KVVRLQ L++ MDNLY DTY+FFVPNRLVW HW+EF GEN Q AW
Sbjct 43 DEVLPGDTFQVKTNKVVRLQPLVSAPMDNLYFDTYYFFVPNRLVWEHWEEFMGENKQGAW 102
Query 62 LPTTEYEIPQITAPATGGWSIGTIADYLGIPTGVPDLSVNALPFRAYALVMNEWFRDENL 121
+P TEY IPQIT+PA+ G+ IGTIADY GIPTGVP+LSV+ALPFRAYAL+++EWFRD+NL
Sbjct 103 IPQTEYTIPQITSPASTGFEIGTIADYFGIPTGVPNLSVSALPFRAYALIVDEWFRDQNL 162
Query 122 TDPLVVPLDDATVAGVNTGAYVTDVAKGGLPFVAAKYHDYFTSCLPAPQKGPDVVIP--- 178
PL +PLDD T+ GVNTG YVTD KGG PFVAAKYHDYFTSCLP+PQKGPDV I
Sbjct 163 QLPLNIPLDDTTLQGVNTGDYVTDTVKGGKPFVAAKYHDYFTSCLPSPQKGPDVTIAAVG 222
Query 179 ---------------------GGTGMSVPVIP--QADKVPSGLITMPYTATFLNETPVRS 215
G + +S + Q + + ++T T + + + +
Sbjct 223 DFPVYTGDPHNNNGSNKALHYGISNISSGSVSFSQGNYIIPSVLTTGSTQSVPAQGKLNA 282
Query 216 TTGIFFNDSGSQTNGVSAGSSEDALPVIDNLWAVGDGVATATINQLRLAFQIQKLYEKDA 275
+ GS + S GS P DNL+A G AT TINQLR+AFQIQKLYEKDA
Sbjct 283 SNITMTTSPGSPDS--SFGSKLSVYP--DNLYA-SSGTAT-TINQLRMAFQIQKLYEKDA 336
Query 276 RGGTRYTEILRSHFGVTSPDSRLQRPEYLGGNRIPIRINQIVQQSATQEGSTPQGNPVGL 335
R G+RY E++RSHF VT D+R+Q PEYLGGNRIPI INQ+VQ S T + S PQGN G
Sbjct 337 RAGSRYRELIRSHFSVTPLDARMQVPEYLGGNRIPININQVVQTSQTSDVS-PQGNVAGQ 395
Query 336 SLTSDNHGDFTKSFTEHGFILGLMVARYDHTYQQGLDRMFSRKSRFDYYWPVFANIGEQA 395
SLTSD+HGDF KSFTEHG ++G+ VARYDHTYQQG+ +++SRK+RFDYYWPV ANIGEQA
Sbjct 396 SLTSDSHGDFIKSFTEHGMLIGVAVARYDHTYQQGVSKLWSRKTRFDYYWPVLANIGEQA 455
Query 396 VLNKEIYAQGTNEDDEVFGYQEAWADYRYKPNRVCGEMRSQAPQSLDVWHLGDDYSKLPS 455
VLNKEIYAQGT +D+EVFGYQEAWA+YRYKP+ V GEMRS A SLD WH DDY+ LP
Sbjct 456 VLNKEIYAQGTAQDEEVFGYQEAWAEYRYKPSIVTGEMRSSARTSLDSWHFADDYNSLPK 515
Query 456 LSDEWIREDKTNVDRVLAVQSSVSNQLFADIYVQNRCTRPMPMYSIPGLIDHH 508
LS +WI+EDKTN+DRVLAV SSVSNQ FAD Y++N TR +P YSIPGLIDHH
Sbjct 516 LSADWIKEDKTNIDRVLAVSSSVSNQYFADFYIENETTRALPFYSIPGLIDHH 568
>gi|575094415|emb|CDL65790.1| unnamed protein product [uncultured bacterium]
Length=569
Score = 520 bits (1339), Expect = 3e-176, Method: Compositional matrix adjust.
Identities = 264/529 (50%), Positives = 337/529 (64%), Gaps = 29/529 (5%)
Query 1 LDEVLPGDTFKIKTSKVVRLQTLITPMMDNLYLDTYFFFVPNRLVWSHWKEFNGENTQSA 60
+DEVLPGDT KIK +VR+ T + P+MDN YLD ++FFVP RLVW HW+ GENT+S
Sbjct 42 VDEVLPGDTIKIKQRSLVRMSTPLYPVMDNCYLDIWYFFVPCRLVWDHWQNLMGENTKSY 101
Query 61 WLPTTEYEIPQITAPATGGWSIGTIADYLGIPTGVPDLSVNALPFRAYALVMNEWFRDEN 120
W P +Y P +AP +GGW +GTIADY+GIPTGV + VN++P RAYA + NEWFRDEN
Sbjct 102 WAPDVQYTTPLTSAP-SGGWQVGTIADYMGIPTGVSGIKVNSMPMRAYARIWNEWFRDEN 160
Query 121 LTDPLVVPLDDATVAGVNTGAYVTDVAKGGLPFVAAKYHDYFTSCLPAPQKGPDVVIP-- 178
L P+ DDAT G NTG +TD GGLP AK+ DYFTSCLPAPQKG +
Sbjct 161 LQQPVTQHSDDATTTGSNTGTELTDAESGGLPLKVAKFKDYFTSCLPAPQKGEAIGFDFN 220
Query 179 -----GGTGMSVPVIPQADKVPSG---------LITMPYTATFLN------ETPVRSTTG 218
G G+ P+ + L+ Y ++ N +T V
Sbjct 221 QTPKVKGIGLVFPLETNTGHTATDILWRQPDAQLVGENYNTSYNNFNSITTQTTVNGKKA 280
Query 219 IFFNDSGSQTNGVSAGSSEDALPVIDN--LWAVGDGVAT-ATINQLRLAFQIQKLYEKDA 275
FFN+ +SA +D ++ L AV + +IN LR A +Q + E DA
Sbjct 281 FFFNNGKGPM--LSARFEDDYNGGVEQVELTAVAENSTNFLSINDLRQAIALQHILEADA 338
Query 276 RGGTRYTEILRSHFGVTSPDSRLQRPEYLGGNRIPIRINQIVQQSATQEGSTPQGNPVGL 335
RGGTRY EIL++ FGV+SPD+RLQR EY+GG RIPI ++Q++Q SA+ + ++PQGN
Sbjct 339 RGGTRYVEILKNEFGVSSPDARLQRSEYIGGERIPINVSQVIQSSAS-DTTSPQGNAAAY 397
Query 336 SLTSDNHGDFTKSFTEHGFILGLMVARYDHTYQQGLDRMFSRKSRFDYYWPVFANIGEQA 395
SLT+ + S EHG+ILGL R DH+YQQGL RM++R RF YY P+ AN+GEQA
Sbjct 398 SLTTSANTIRAYSAVEHGYILGLAAIRVDHSYQQGLSRMWTRSDRFSYYHPMLANLGEQA 457
Query 396 VLNKEIYAQGTNEDDEVFGYQEAWADYRYKPNRVCGEMRSQAPQSLDVWHLGDDYSKLPS 455
VLN+EIYAQGT D EVFGYQEAWADYRY+ N + GEMRS QSLD WH GD Y+ LP
Sbjct 458 VLNQEIYAQGTTADTEVFGYQEAWADYRYRTNMITGEMRSTYAQSLDAWHYGDKYTDLPR 517
Query 456 LSDEWIREDKTNVDRVLAVQSSVSNQLFADIYVQNRCTRPMPMYSIPGL 504
LS++WI+E + N+DR LAVQS S+Q ++Y RPMP+YS+PGL
Sbjct 518 LSNDWIKEGQENIDRTLAVQSENSHQFICNLYFDQTWVRPMPIYSVPGL 566
>gi|575094431|emb|CDL65804.1| unnamed protein product [uncultured bacterium]
Length=560
Score = 503 bits (1296), Expect = 8e-170, Method: Compositional matrix adjust.
Identities = 265/528 (50%), Positives = 340/528 (64%), Gaps = 29/528 (5%)
Query 1 LDEVLPGDTFKIKTSKVVRLQTLITPMMDNLYLDTYFFFVPNRLVWSHWKEFNGENTQSA 60
+DEVLPGDTF++ + ++R T I P+MDN +LD YFFFVPNRL W HW+E GEN +A
Sbjct 42 VDEVLPGDTFELDMTAIIRGSTPIFPVMDNSFLDVYFFFVPNRLTWEHWRELMGENRTTA 101
Query 61 WLPTTEYEIPQITAPATGGWSIGTIADYLGIPTGVPDLSVNALPFRAYALVMNEWFRDEN 120
W +Y +PQ+TAPA GGW ++AD++GIPT V ++SVNALPFRAY L+ NE+FR++N
Sbjct 102 WTQPVDYSVPQVTAPA-GGWEELSLADHMGIPTKVDNISVNALPFRAYGLIYNEFFRNQN 160
Query 121 LTDPLVVPLDDATVAGVNTGAYVTD---VAKGGLPFVAAKYHDYFTSCLPAPQKGPDVVI 177
LT+P V + DA +AG N G +AK+ DYFT LP PQKG V I
Sbjct 161 LTNPTQVEVTDANIAGKNPNDVKNSNDWAITGAKCLKSAKFFDYFTGALPQPQKGEPVEI 220
Query 178 -------PGGTG-MSVPVIPQADKVPSGLITMPYTATFLNETPVRSTTGIFFNDSGSQTN 229
P G G P+ DKV + + + T G+ + N
Sbjct 221 NLASSWLPVGIGDYHGPL----DKVSNSDTLTWESPSSEGNTKRTYALGMVQQEGEVNPN 276
Query 230 GV------SAGSSEDALPVI---DNLWAVGDGVATATINQLRLAFQIQKLYEKDARGGTR 280
G+ + GS ++ V NLWA A AT+NQLR AFQ+QKL EKDARGGTR
Sbjct 277 GLKNFETKAGGSFSESGAVAAYPTNLWA-SPVTAAATVNQLRQAFQVQKLLEKDARGGTR 335
Query 281 YTEILRSHFGVTSPDSRLQRPEYLGGNRIPIRINQIVQQSATQEGSTPQGNPVGLSLTSD 340
Y EIL++HFGVT+ D+R+Q PEYLGG ++PI ++Q+VQ SA+ + S PQGN +S+T
Sbjct 336 YREILKNHFGVTTSDARMQIPEYLGGCKVPINVSQVVQTSASTDAS-PQGNTAAISVTPF 394
Query 341 NHGDFTKSFTEHGFILGLMVARYDHTYQQGLDRMFSRKSRFDYYWPVFANIGEQAVLNKE 400
+ FTKSF EHGFI+G+ AR +YQQG++RM+SRK R DYY+PV ANIGEQA+LNKE
Sbjct 395 SKSMFTKSFDEHGFIIGVATARTAQSYQQGIERMWSRKDRLDYYFPVLANIGEQAILNKE 454
Query 401 IYAQGTNEDDEVFGYQEAWADYRYKPNRVCGEMRSQAPQSLDVWHLGDDYSKLPSLSDEW 460
IYAQG +DDE FGYQEAWADYRYKPN +CG RS A QSLD WH G DY KLP+LS +W
Sbjct 455 IYAQGNAKDDEAFGYQEAWADYRYKPNTICGRFRSNAQQSLDAWHYGQDYDKLPTLSTDW 514
Query 461 IREDKTNVDRVLAVQSSVSNQLFADIYVQNRCTRPMPMYSIPGLIDHH 508
+ + + R LAVQ+ A+ + R MP+YSIPGLIDH+
Sbjct 515 MEQSDIEMKRTLAVQTEPD--FIANFRFNCKTVRVMPLYSIPGLIDHN 560
>gi|530695385|gb|AGT39938.1| major capsid protein [Marine gokushovirus]
Length=514
Score = 436 bits (1120), Expect = 4e-144, Method: Compositional matrix adjust.
Identities = 248/509 (49%), Positives = 315/509 (62%), Gaps = 48/509 (9%)
Query 2 DEVLPGDTFKIKTSKVVRLQTLITPMMDNLYLDTYFFFVPNRLVWSHWKEFNGENTQSAW 61
DE LPGDTF + + RL T I P MDNLY++T+FF VP RL+W++W++F GE
Sbjct 50 DEALPGDTFTMDANGFGRLATPIAPFMDNLYIETFFFAVPYRLIWTNWEKFCGEQDNPG- 108
Query 62 LPTTEYEIPQITAPATGGWSIGTIADYLGIPTGVPDLSVNALPFRAYALVMNEWFRDENL 121
+T+Y +PQ TG S T+ DY G+PT V +L+ N L RAY LV NEWFRD+NL
Sbjct 109 -DSTDYLVPQ----TTGTISNSTLYDYFGVPTDV-NLTFNNLCGRAYNLVYNEWFRDQNL 162
Query 122 TDPLVVPLDDATVAGVNTGAYVTDVAKGGLPFVAAKYHDYFTSCLPAPQKGPDVVIPGGT 181
+ + V D G +T + T + +G K HDYFTS LP PQKG V +P GT
Sbjct 163 QNSVTVDKGD----GPDTASNYTLLKRG-------KRHDYFTSALPWPQKGEAVTLPLGT 211
Query 182 GMSVPVIPQADKVPSGLITMP--YTATFLNETPVRSTTGIF-FNDSGSQTNGVSAGSSED 238
+ P++ T P Y + N P + G + F +G G+
Sbjct 212 --TAPIMS------GDFTTTPTNYIPSNGNNIPPQDANGDYSFAGTGVGGYGI------- 256
Query 239 ALPVIDNLWAVGDGVATATINQLRLAFQIQKLYEKDARGGTRYTEILRSHFGVTSPDSRL 298
WA ATINQLR AFQIQ+LYEKDARGGTRYTE+++SHFGVTSPD+RL
Sbjct 257 --------WADLSDATAATINQLREAFQIQRLYEKDARGGTRYTEVIQSHFGVTSPDARL 308
Query 299 QRPEYLGGNRIPIRINQIVQQSATQEGSTPQGNPVGLSLTSDNHGDFTKSFTEHGFILGL 358
QRPEYLGG + I IN I Q S+T + +TPQGN G T F KSFTEH +LGL
Sbjct 309 QRPEYLGGGKDRININPIAQTSST-DATTPQGNLSGYGTTGFTGHRFNKSFTEHSVVLGL 367
Query 359 MVARYDHTYQQGLDRMFSRKSRFDYYWPVFANIGEQAVLNKEIYAQGTNEDDEVFGYQEA 418
D TYQQGL R FSR++R+D+YWP A++GEQAVLNKEIYAQGT +D+ VFGYQE
Sbjct 368 ACVFADLTYQQGLPRHFSRQTRWDFYWPALAHLGEQAVLNKEIYAQGTTDDNNVFGYQER 427
Query 419 WADYRYKPNRVCGEMRSQAPQSLDVWHLGDDYSKLPSLSDEWIREDKTNVDRVLAVQSSV 478
+A+YRYKP+ + G+MRS QSLD+WHL D+ LP L+ +I E+ VDRV AVQ+
Sbjct 428 YAEYRYKPSSITGQMRSNFAQSLDIWHLAQDFGSLPVLNSSFIEENPP-VDRVTAVQNYP 486
Query 479 SNQLFADIYVQNRCTRPMPMYSIPGLIDH 507
+ L D+Y + +C RPMP Y +PGLIDH
Sbjct 487 N--LILDMYFKLKCARPMPTYGVPGLIDH 513
>gi|444297960|dbj|GAC77859.1| major capsid protein, partial [uncultured marine virus]
Length=494
Score = 429 bits (1103), Expect = 1e-141, Method: Compositional matrix adjust.
Identities = 230/510 (45%), Positives = 311/510 (61%), Gaps = 25/510 (5%)
Query 1 LDEVLPGDTFKIKTSKVVRLQTLITPMMDNLYLDTYFFFVPNRLVWSHWKEFNGENTQSA 60
+DE LPGDTF + ++ R+ T I P+MDNL +D++FF VP RL+W +W +GE
Sbjct 6 VDEALPGDTFSVSSTFFARMATPIFPIMDNLKMDSFFFAVPVRLLWDNWARMHGEQRNPG 65
Query 61 WLPTTEYEIPQITAPATGGWSIGTIADYLGIPTGVPDLSVNALPFRAYALVMNEWFRDEN 120
+T++ +P +T+P G+ G++ DYLG+PTG+PDL ++L RA+ L+ NEWFRDEN
Sbjct 66 --DSTDFVVPTMTSPPINGYDEGSLEDYLGLPTGIPDLEHSSLFHRAHNLIHNEWFRDEN 123
Query 121 LTDPLVVPLDDATVAGVNTGAYVTDVAKGGLPFVAAKYHDYFTSCLPAPQKGPDVVIPGG 180
LTD ++ +DD + ++ Y +K HDYFTS LP PQKG + IP G
Sbjct 124 LTDSVINNVDDGPDSNLDYALYRR-----------SKRHDYFTSALPWPQKGESISIPLG 172
Query 181 TGMSVPVIPQADKVPSGLITMPYTATFLNETPVRSTTGIFFNDSGSQTNGVSAGSSEDAL 240
T V I + D+ G Y + + S T I G + G + ED
Sbjct 173 TRADVKGIGKEDQT-FGASVNAYESGGTGQVQYLSATRI-----GDGSAGETHSMEEDPN 226
Query 241 -PVIDNLWAVGDGVATATINQLRLAFQIQKLYEKDARGGTRYTEILRSHFGVTSPDSRLQ 299
P N++A ATINQLR +FQIQK+ E+DARGGTR TE++ +HFGV SPD+R+Q
Sbjct 227 NPGFPNIYADLTTATAATINQLRQSFQIQKMLERDARGGTRLTEVILAHFGVRSPDARMQ 286
Query 300 RPEYLGGNRIPIRINQIVQQSATQ--EGSTPQGNPVGLSLTSDNHGDFTKSFTEHGFILG 357
RPEYLGG PI + Q+ E +TPQGN + ++ FTKSFTEH ILG
Sbjct 287 RPEYLGGGSAPIALQQVASTVPNDFTENNTPQGNLAAYGIGVSSNNSFTKSFTEHCIILG 346
Query 358 LMVARYDHTYQQGLDRMFSRKSRFDYYWPVFANIGEQAVLNKEIYAQGTNEDDEVFGYQE 417
+ R D TYQQGL+RMFSR +R+D+Y+P ++IGEQAVLNKEIYAQG D++VFGYQE
Sbjct 347 YVNVRADITYQQGLNRMFSRSTRYDFYYPALSHIGEQAVLNKEIYAQGLPADEDVFGYQE 406
Query 418 AWADYRYKPNRVCGEMRSQAPQSLDVWHLGDDYSKLPSLSDEWIREDKTNVDRVLAVQSS 477
A+YRYKP+++ G RS A LD WHL D++ LP L +I E+ +DRV+AV +
Sbjct 407 RHAEYRYKPSQISGAFRSSAAAPLDAWHLSQDFATLPVLDQTFIEENPP-IDRVIAVPTE 465
Query 478 VSNQLFADIYVQNRCTRPMPMYSIPGLIDH 507
D Y +C RPMP+Y +PGLIDH
Sbjct 466 P--HFLFDSYTSMKCARPMPVYGVPGLIDH 493
>gi|313766927|gb|ADR80653.1| putative major coat protein [Uncultured Microviridae]
Length=533
Score = 430 bits (1106), Expect = 1e-141, Method: Compositional matrix adjust.
Identities = 235/507 (46%), Positives = 319/507 (63%), Gaps = 39/507 (8%)
Query 1 LDEVLPGDTFKIKTSKVVRLQTLITPMMDNLYLDTYFFFVPNRLVWSHWKEFNGENTQSA 60
+DEVLPGDTF++ + RL T + P+MDN+Y++T+FF+VPNR++W +W++FNG Q
Sbjct 50 VDEVLPGDTFQMNATGFGRLATPLYPVMDNMYVETFFFYVPNRIIWDNWEKFNG--AQDD 107
Query 61 WLPTTEYEIPQITAPATGGWSIGTIADYLGIPTGVPDLSVNALPFRAYALVMNEWFRDEN 120
+T++ +PQI + + G++ DY+G+PT + + N L RAY L+ NEWFRDEN
Sbjct 108 PNDSTDFLVPQIQSATV---AEGSLFDYMGLPTQIAGIDFNNLHGRAYNLIWNEWFRDEN 164
Query 121 LTDPLVVPLDDATVAGVNTGAYVTDVAKGGLPFVAAKYHDYFTSCLPAPQKGPDVVIPGG 180
L D L VP DD G +T T +G K HDYFTS LP PQKG V +P G
Sbjct 165 LQDSLGVPKDD----GPDTYTGYTIQKRG-------KRHDYFTSALPWPQKGDAVSLPLG 213
Query 181 TGMSVPVIPQADKVPSGLITMPYTATFLNETPVRSTTGIFFNDSGSQTNGVSAGSSEDAL 240
T + A L PV +S G+
Sbjct 214 TSADIHTAAAAGTDIGIYSVGSSDFRLLTSDPVEV--------------ALSGGTP---- 255
Query 241 PVIDNLWAVGDGVATATINQLRLAFQIQKLYEKDARGGTRYTEILRSHFGVTSPDSRLQR 300
P + ++A ATINQLR AFQIQ+LYEKDARGGTRYTEIL+SHFGVTSPD+RLQR
Sbjct 256 PETNKMFADLSNATAATINQLREAFQIQRLYEKDARGGTRYTEILQSHFGVTSPDARLQR 315
Query 301 PEYLGGNRIPIRINQIVQQSATQEGSTPQGNPVGLSLTSDNHGDFTKSFTEHGFILGLMV 360
PEYLGG + + + Q V Q+++ + ++PQGN L T+ + G F+KSF EHG ++GL
Sbjct 316 PEYLGGQKTEVMM-QTVPQTSSTDSTSPQGNLAALG-TATSRGGFSKSFVEHGVLIGLAC 373
Query 361 ARYDHTYQQGLDRMFSRKSRFDYYWPVFANIGEQAVLNKEIYAQGTNEDDEVFGYQEAWA 420
D TYQQG++RM+SR+ R+D+YWP A++GEQAVLN+EIY QGT+ D + FGYQE +A
Sbjct 374 VFADLTYQQGMNRMWSRRDRWDFYWPSLAHLGEQAVLNQEIYTQGTSADTQTFGYQERFA 433
Query 421 DYRYKPNRVCGEMRSQAPQSLDVWHLGDDYSKLPSLSDEWIREDKTNVDRVLAVQSSVSN 480
+YRYKP+++ G+MRS A +LD WHL D++ LP+L+ +I E+ VDRV+AV S
Sbjct 434 EYRYKPSQITGKMRSNATGTLDAWHLAQDFTALPALNASFIEENPP-VDRVIAVPS--EP 490
Query 481 QLFADIYVQNRCTRPMPMYSIPGLIDH 507
+ D Y + TRPMP+YS+PGLIDH
Sbjct 491 EFIWDWYFDLKTTRPMPVYSVPGLIDH 517
Lambda K H a alpha
0.318 0.136 0.417 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 3600440468988