bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: All non-redundant GenBank CDS translations+PDB+SwissProt+PIR+PRF
excluding environmental samples from WGS projects
49,011,213 sequences; 17,563,301,199 total letters
Query= Contig-2_CDS_annotation_glimmer3.pl_2_4
Length=636
Score E
Sequences producing significant alignments: (Bits) Value
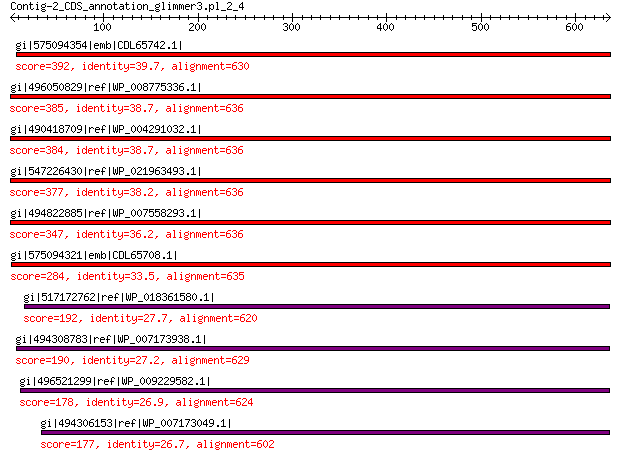
gi|575094354|emb|CDL65742.1| unnamed protein product 392 6e-124
gi|496050829|ref|WP_008775336.1| hypothetical protein 385 1e-121
gi|490418709|ref|WP_004291032.1| hypothetical protein 384 4e-121
gi|547226430|ref|WP_021963493.1| putative uncharacterized protein 377 7e-119
gi|494822885|ref|WP_007558293.1| hypothetical protein 347 9e-107
gi|575094321|emb|CDL65708.1| unnamed protein product 284 1e-82
gi|517172762|ref|WP_018361580.1| hypothetical protein 192 9e-50
gi|494308783|ref|WP_007173938.1| hypothetical protein 190 4e-49
gi|496521299|ref|WP_009229582.1| capsid protein 178 6e-45
gi|494306153|ref|WP_007173049.1| hypothetical protein 177 8e-45
>gi|575094354|emb|CDL65742.1| unnamed protein product [uncultured bacterium]
Length=615
Score = 392 bits (1007), Expect = 6e-124, Method: Compositional matrix adjust.
Identities = 250/670 (37%), Positives = 361/670 (54%), Gaps = 97/670 (14%)
Query 7 LKELQNHPHKAGFDIGSKNLFTAKVGELLPVYWDFAIPSCDYDIDLAYFTRTRPVQTAAY 66
+ +++N P + GFD+ K FTAK GELLPV +P ++I+L FTRT+P+ T+A+
Sbjct 3 MADIKNRPSRNGFDLSFKKNFTAKAGELLPVMTKVVLPGDSFNINLRSFTRTQPLNTSAF 62
Query 67 TRIREYFDFYAVPCDLLWKSFDSAVIQM-GEVAPVQAKTPLDPLTVGTDIPWCTLSDLYT 125
R+REY+DFY VP + +W FDS + QM V T D + +P+ T +
Sbjct 63 ARMREYYDFYFVPFEQMWNKFDSCITQMNANVQHASGPTLDDNTPLSGRMPYFTSEQIAD 122
Query 126 SLIFMNNRVSLGQTVSVVPDYSNIFGYSRCDTSHKLLLYLNYGNFVEPSSSNVGTSTNRW 185
++N++ + + N FG++R + KLL YL YG++ ++ + TN W
Sbjct 123 ---YLNDQATAAR--------KNPFGFNRSTLTCKLLQYLGYGDY-----NSFDSETNTW 166
Query 186 FNTSFTSSAVQNYSQKYSQNVSVSLFPLLAYQKIYQDFFRWSQWENADPTAYNVDYYNGS 245
++ N+ +S FPLLAYQKIY DF+R++QWE +P+ +N+DY G+
Sbjct 167 ------------SAKPLLYNLELSPFPLLAYQKIYSDFYRYTQWEKTNPSTFNLDYIKGT 214
Query 246 GNLFGNGGIASSIPSSNDYWKRDNMFSLRYCNWNKDMFMGLLPNSQFGDVAVVSGVEGID 305
+L + + +PS + +N F +RYCN+ KDMF G+LP +Q+G +V
Sbjct 215 SDLQMD---LTGLPSDD-----NNFFDIRYCNYQKDMFHGVLPVAQYGSASV-------- 258
Query 306 TFVPVE-VFNSINETNIAKPPLTGTHTPVYTDDAMTSSTTPSRIRVAGGSGIPS---SVA 361
VP+ N I +N P+ T TP D T T S + V G G+ + V+
Sbjct 259 --VPINGQLNVI--SNGDSGPIFKTSTP----DPGTPGT--SYVTVGGNIGVDNRSFGVS 308
Query 362 GSALGV-----------------RSLL------------GGEFSILALRQAEALQKWKEI 392
GS L V RSLL G ILALRQAE LQKWKE+
Sbjct 309 GSTLNVGKSADPSGYGFPSNASTRSLLWENPNLIIENNQGFYVPILALRQAEFLQKWKEV 368
Query 393 TQSVDTNYRDQIKAHFGINTPASMSHMAQYIGGIARNLDISEVVNNNLSETGSEAV-IYG 451
+ S + +Y+ QI+ H+GI +SH A+Y+GG A +LDI+EV+NNN+ TG A I G
Sbjct 369 SVSGEEDYKSQIEKHWGIKVSDFLSHQARYLGGCATSLDINEVINNNI--TGDNAADIAG 426
Query 452 KGVGTGSGKMRYHTGSQYCIIMCIYHAVPLLDYAISGQDSQLLCTSVEDLPIPEFDNIGM 511
KG TG+G +R+ + +Y IIMCIYH +P++DY SG D PIPE D IGM
Sbjct 427 KGTFTGNGSIRFESKGEYGIIMCIYHVLPIVDYVGSGVDHSCTLVDATSFPIPELDQIGM 486
Query 512 EAVPAITLFNSNAFDNDLESDFDFLGYNPRYWPWKSKIDRVHGAFLTTLKDWVAPIDDFY 571
E+VP + N ++D S FLGY PRY WK+ +DR G F +L+ W P+ D
Sbjct 487 ESVPLVRAMNP-VKESDTPSADTFLGYAPRYIDWKTSVDRSVGDFADSLRTWCLPVGDKE 545
Query 572 LNRW----FASGGSSQA-SISWPFFKVNPNTLDSIFAVAADSTWESDQLLINCDVSCKVV 626
L F S + + SI+ FFKVNP+ +D +FAV ADST ++D+ L + KVV
Sbjct 546 LTSANSLNFPSNPNVEPDSIAAGFFKVNPSIVDPLFAVVADSTVKTDEFLCSSFFDVKVV 605
Query 627 RPLSQDGMPY 636
R L +G+PY
Sbjct 606 RNLDVNGLPY 615
>gi|496050829|ref|WP_008775336.1| hypothetical protein [Bacteroides sp. 2_2_4]
gi|229448893|gb|EEO54684.1| putative capsid protein (F protein) [Bacteroides sp. 2_2_4]
Length=580
Score = 385 bits (988), Expect = 1e-121, Method: Compositional matrix adjust.
Identities = 246/648 (38%), Positives = 350/648 (54%), Gaps = 80/648 (12%)
Query 1 MAHFTGLKELQNHPHKAGFDIGSKNLFTAKVGELLPVYWDFAIPSCDYDIDLAYFTRTRP 60
MA+ LK L+N + GFD+ SK FTAK GELLPV +P + IDL FTRT+P
Sbjct 1 MANIMSLKSLRNKTSRNGFDLSSKRNFTAKPGELLPVKCWEVLPGDKWSIDLKSFTRTQP 60
Query 61 VQTAAYTRIREYFDFYAVPCDLLWKSFDSAVIQMGEVAPVQAKT--PLDPLTVGTDIPWC 118
+ TAA+ R+REY+DFY VP +LLW ++ + QM + P A + P + +P
Sbjct 61 LNTAAFARMREYYDFYFVPYNLLWNKANTVLTQMYD-NPQHATSYIPSANQALAGVMPNV 119
Query 119 TLSDLYTSLIFMNNRVSLGQTVSVVPDYSNIFGYSRCDTSHKLLLYLNYGNFVEPSSSNV 178
T + L + V+ + N FGYSR + KLL YL YGNF ++S
Sbjct 120 TCKGIADYLNLVAPDVTTTNSYE-----KNYFGYSRSLGTAKLLEYLGYGNFYTYATSK- 173
Query 179 GTSTNRWFNTSFTSSAVQNYSQKYSQNVSVSLFPLLAYQKIYQDFFRWSQWENADPTAYN 238
N ++T S + S N+ ++++ +LAYQKIY D R SQWE P+ +N
Sbjct 174 --------NNTWTKSPL-------SSNLQLNIYGVLAYQKIYADHIRDSQWEKVSPSCFN 218
Query 239 VDYYNGSGNLFGNGGIASSIPSSNDYWKRDNMFSLRYCNWNKDMFMGLLPNSQFGDVAVV 298
VDY +G+ + S+ + + NMF LRYCNW KD+F G+LP Q+GD A V
Sbjct 219 VDYLSGT---VDSAMTIDSMITGQGFAPFYNMFDLRYCNWQKDLFHGVLPRQQYGDTAAV 275
Query 299 SGVEGIDTFVPVEVFNSINETNIAKPPLTGTHTPVYTDDAMTSSTTPSRIRVAGGSGIPS 358
++N +N+ A TP V G S
Sbjct 276 ----------------NVNLSNVLS--------------AQYMVQTPDGDPVGG-----S 300
Query 359 SVAGSALGVRSLLG-GEFSILALRQAEALQKWKEITQSVDTNYRDQIKAHFGINTPASMS 417
+ + + ++++ G G F++LALRQAE LQKWKEITQS + +Y+DQI+ H+ ++ + S
Sbjct 301 PFSSTGVNLQTVNGSGTFTVLALRQAEFLQKWKEITQSGNKDYKDQIEKHWNVSVGEAYS 360
Query 418 HMAQYIGGIARNLDISEVVNNNLSETGSEAV-IYGKGVGTGSGKMRYHTGSQYCIIMCIY 476
M+ Y+GG +LDI+EVVNNN+ TGS A I GKGV G+G++ + G +Y +IMCIY
Sbjct 361 EMSLYLGGTTASLDINEVVNNNI--TGSNAADIAGKGVVVGNGRISFDAGERYGLIMCIY 418
Query 477 HAVPLLDYAISGQDSQLLCTSVEDLPIPEFDNIGMEAVPAITLFNSNAFDNDLESDFD-- 534
H++PLLDY + + D IPEFD +GME+VP ++L N L+S ++
Sbjct 419 HSLPLLDYTTDLVNPAFTKINSTDFAIPEFDRVGMESVPLVSLMNP------LQSSYNVG 472
Query 535 --FLGYNPRYWPWKSKIDRVHGAFLTTLKDWVAPIDDF----YLNRWFASGGSSQASISW 588
LGY PRY +K+ +D GAF TTLK WV D+ LN S +++
Sbjct 473 SSILGYAPRYISYKTDVDSSVGAFKTTLKSWVMSYDNQSVINQLNYQDDPNNSPGTLVNY 532
Query 589 PFFKVNPNTLDSIFAVAADSTWESDQLLINCDVSCKVVRPLSQDGMPY 636
FKVNPN +D +FAVAA ++ ++DQ L + KVVR L DG+PY
Sbjct 533 TNFKVNPNCVDPLFAVAASNSIDTDQFLCSSFFDVKVVRNLDTDGLPY 580
>gi|490418709|ref|WP_004291032.1| hypothetical protein [Bacteroides eggerthii]
gi|217986636|gb|EEC52970.1| putative capsid protein (F protein) [Bacteroides eggerthii DSM
20697]
Length=578
Score = 384 bits (985), Expect = 4e-121, Method: Compositional matrix adjust.
Identities = 246/653 (38%), Positives = 346/653 (53%), Gaps = 92/653 (14%)
Query 1 MAHFTGLKELQNHPHKAGFDIGSKNLFTAKVGELLPVYWDFAIPSCDYDIDLAYFTRTRP 60
MA+ LK ++N P + GFD+ K FTAK GELLPV +P + I+L FTRT+P
Sbjct 1 MANIMSLKSIRNKPSRNGFDLSFKKNFTAKAGELLPVMVKEVLPGDTFKINLKAFTRTQP 60
Query 61 VQTAAYTRIREYFDFYAVPCDLLWKSFDSAVIQMGEVAPVQAKTPLDP---LTVGTDIPW 117
V TAA+ RIREY+DF+ VP DLLW ++ + QM + Q +DP + ++P+
Sbjct 61 VNTAAFARIREYYDFFFVPYDLLWNKANTVLTQMYDNP--QHAVSIDPTRNFVLSGEMPY 118
Query 118 CTLSDLYTSLIFMNNRVSLGQTVSVVPDY-SNIFGYSRCDTSHKLLLYLNYGNFVEPSSS 176
T S+ S I N +S T S + DY SN FGY+R +S KLL YL YGN+
Sbjct 119 MT-SEAIASYI---NALS---TASALADYKSNYFGYNRSKSSVKLLEYLGYGNYES---- 167
Query 177 NVGTSTNRWFNTSFTSSAVQNYSQKYSQNVSVSLFPLLAYQKIYQDFFRWSQWENADPTA 236
T+ W + N++ ++F LLAYQKIY DF+R SQWE P+
Sbjct 168 ---FLTDDWNTAPLMA------------NLNHNIFGLLAYQKIYSDFYRDSQWERVSPST 212
Query 237 YNVDYYNGSGNLFGNGGIASSIPSSNDYWKRDNMFSLRYCNWNKDMFMGLLPNSQFGDVA 296
+NVDY +GS N S ++++ N F LRYCNW KD+F G+LP+ Q+G+ A
Sbjct 213 FNVDYLDGSSMNLDNA-------YSTEFYQNYNFFDLRYCNWQKDLFHGVLPHQQYGETA 265
Query 297 VVSGVEGIDTFVPVEVFNSINETNIAKPPLTGTHTPVYTDDAMTSSTTPSRIRVAGGSGI 356
V S P +TG T TS TT A G+
Sbjct 266 VAS----------------------ITPDVTGKLTLSNFSTVGTSPTT------ASGTAT 297
Query 357 PSSVAGSALGVRSLLGGEFSILALRQAEALQKWKEITQSVDTNYRDQIKAHFGINTPASM 416
+ A + G+ SIL LRQAE LQKWKEITQS + +Y+DQ++ H+G++
Sbjct 298 KNLPAFDTV-------GDLSILVLRQAEFLQKWKEITQSGNKDYKDQLEKHWGVSVGDGF 350
Query 417 SHMAQYIGGIARNLDISEVVNNNLSETGSEAV-IYGKGVGTGSGKMRYHTGSQYCIIMCI 475
S + Y+GG++ ++DI+EV+N N+ TGS A I GKGVG +G++ +++ +Y +IMCI
Sbjct 351 SELCTYLGGVSSSIDINEVINTNI--TGSAAADIAGKGVGVANGEINFNSNGRYGLIMCI 408
Query 476 YHAVPLLDYAISGQDSQLLCTSVEDLPIPEFDNIGMEAVPAITLFNS-NAFDNDLESDFD 534
YH +PLLDY D L + D IPEFD +GM+++P + L N +F N +
Sbjct 409 YHCLPLLDYTTDMLDPAFLKVNSTDYAIPEFDRVGMQSMPLVQLMNPLRSFAN---ASGL 465
Query 535 FLGYNPRYWPWKSKIDRVHGAFLTTLKDWVAPIDDFYLNRWF-----------ASGGSSQ 583
LGY PRY +K+ +D+ G F TL WV + + + + S
Sbjct 466 VLGYVPRYIDYKTSVDQSVGGFKRTLNSWVISYGNISVLKQVTLPNDAPPIEPSEPVPSV 525
Query 584 ASISWPFFKVNPNTLDSIFAVAADSTWESDQLLINCDVSCKVVRPLSQDGMPY 636
A +++ FFKVNP+ LD IFAV A +DQ L + K VR L DG+PY
Sbjct 526 APMNFTFFKVNPDCLDPIFAVQAGDDTNTDQFLCSSFFDIKAVRNLDTDGLPY 578
>gi|547226430|ref|WP_021963493.1| putative uncharacterized protein [Prevotella sp. CAG:1185]
gi|524103382|emb|CCY83994.1| putative uncharacterized protein [Prevotella sp. CAG:1185]
Length=573
Score = 377 bits (968), Expect = 7e-119, Method: Compositional matrix adjust.
Identities = 243/643 (38%), Positives = 348/643 (54%), Gaps = 77/643 (12%)
Query 1 MAHFTGLKELQNHPHKAGFDIGSKNLFTAKVGELLPVYWDFAIPSCDYDIDLAYFTRTRP 60
M+ L L+N + GFD+ KN FTAKVGELLP+ P ++I FTRT+P
Sbjct 1 MSSVMSLTALKNSVKRNGFDLSFKNAFTAKVGELLPIMCKEVYPGDKFNIRGQAFTRTQP 60
Query 61 VQTAAYTRIREYFDFYAVPCDLLWKSFDSAVIQMGEVAPVQAKTPLDPLTVGTDIPWCTL 120
V +AAY+R+REY+DFY VP LLW + M + P A + + + PW T
Sbjct 61 VNSAAYSRLREYYDFYFVPYRLLWNMAPTFFTNMPD--PHHAADLVSSVNLSQRHPWFTF 118
Query 121 SDLYTSLIFMNNRVSLGQTVSVVPDYSNIFGYSRCDTSHKLLLYLNYGNFVEPSSSNVGT 180
D+ + ++ N SL N FG+SR + S KLL YLNYG
Sbjct 119 FDI---MEYLGNLNSLSGAYEKYQ--KNFFGFSRVELSVKLLNYLNYG------------ 161
Query 181 STNRWFNTSFTSSAVQNYSQKYSQNVSVSLFPLLAYQKIYQDFFRWSQWENADPTAYNVD 240
F + S V + S ++ +S FPLLAYQKI +D+FR QW++A P YN+D
Sbjct 162 -----FGKDYESVKVPS----DSDDIVLSPFPLLAYQKICEDYFRDDQWQSAAPYRYNLD 212
Query 241 YYNGSGNLFGNGGIASSIPSS---NDYWKRDNMFSLRYCNWNKDMFMGLLPNSQFGDVAV 297
Y G + F IP S ND +K MF L YCN+ KD F G+LP +Q+GDV+V
Sbjct 213 YLYGKSSGF-------HIPMSSFTNDAFKNPTMFDLNYCNFQKDYFTGMLPRAQYGDVSV 265
Query 298 VSGVEGIDTFVPVEVFNSINETNIAKPPLTGTHTPVYTDDAMTSSTTPSRIRVAGGSGIP 357
S + G +++ +S + T A P G +T + G +
Sbjct 266 ASPIFG-----DLDIGDSSSLT-FASAPQQGANT------------------IQSGVLVV 301
Query 358 SSVAGSALGVRSLLGGEFSILALRQAEALQKWKEITQSVDTNYRDQIKAHFGINTPASMS 417
++ + + G+ S+LALRQAE LQKW+EI QS +Y+ Q++ HF ++ A++S
Sbjct 302 NNNSNTTAGL--------SVLALRQAECLQKWREIAQSGKMDYQTQMQKHFNVSPSATLS 353
Query 418 HMAQYIGGIARNLDISEVVNNNLSETGSEAVIYGKGVGTGSGKMRYHTGSQYCIIMCIYH 477
+Y+GG NLDISEVVN NL+ ++A I GKG GT +G S++ IIMCIYH
Sbjct 354 GHCKYLGGWTSNLDISEVVNTNLT-GDNQADIQGKGTGTLNGNKVDFESSEHGIIMCIYH 412
Query 478 AVPLLDYAISGQDSQLLCTSVEDLPIPEFDNIGMEAV-PAITLFNSNAFDNDLESDFDFL 536
+PLLD++I+ Q T+ D IPEFD++GM+ + P+ +F +D S +
Sbjct 413 CLPLLDWSINRIARQNFKTTFTDYAIPEFDSVGMQQLYPSEMIFGLEDLPSDPSS--INM 470
Query 537 GYNPRYWPWKSKIDRVHGAFLTTLKDWVAPIDDFYLNRWFAS---GGSSQASISWPFFKV 593
GY PRY K+ ID +HG+F+ TL WV+P+ D Y++ + + G S ++++ FFKV
Sbjct 471 GYVPRYADLKTSIDEIHGSFIDTLVSWVSPLTDSYISAYRQACKDAGFSDITMTYNFFKV 530
Query 594 NPNTLDSIFAVAADSTWESDQLLINCDVSCKVVRPLSQDGMPY 636
NP+ +D+IF V ADST +DQLLIN K VR +G+PY
Sbjct 531 NPHIVDNIFGVKADSTINTDQLLINSYFDIKAVRNFDYNGLPY 573
>gi|494822885|ref|WP_007558293.1| hypothetical protein [Bacteroides plebeius]
gi|198272099|gb|EDY96368.1| putative capsid protein (F protein) [Bacteroides plebeius DSM
17135]
Length=613
Score = 347 bits (890), Expect = 9e-107, Method: Compositional matrix adjust.
Identities = 230/655 (35%), Positives = 342/655 (52%), Gaps = 68/655 (10%)
Query 1 MAHFTGLKELQNHPHKAGFDIGSKNLFTAKVGELLPVYWDFAIPSCDYDIDLAYFTRTRP 60
MA+ +K ++N P +AG+D+ K FTAK G L+PV+W +P D + + F RT+P
Sbjct 8 MANIMSMKSVRNKPTRAGYDLTQKINFTAKAGSLIPVWWTPVLPFDDLNATVKSFVRTQP 67
Query 61 VQTAAYTRIREYFDFYAVPCDLLWKSFDSAVIQMGEVAPVQAKTPL--DPLTVGTDIPWC 118
+ TAA+ R+R YFDFY VP +W F +A+ QM + A P+ D + + ++P+
Sbjct 68 LNTAAFARMRGYFDFYFVPFRQMWNKFPTAITQM-RTNLLHASGPVLADNVPLSDELPYF 126
Query 119 TLSDLYTSLIFMNNRVSLGQTVSVVPDYSNIFGYSRCDTSHKLLLYLNYGNFVEPSSSNV 178
T + + VSL D N FGY R +L YL YG+F
Sbjct 127 TAEQVADYI------VSLA-------DSKNQFGYYRAWLVCIILEYLGYGDFYPYIVEAA 173
Query 179 GTSTNRWFNTSFTSSAVQNYSQKYSQNVSVSLFPLLAYQKIYQDFFRWSQWENADPTAYN 238
G W ++ N+ S FPL AYQKIY DF R++QWE ++P+ +N
Sbjct 174 GGEGATW------------ATRPMLNNLKFSPFPLFAYQKIYADFNRYTQWERSNPSTFN 221
Query 239 VDYYNGSGNLFGNGGIASSIPSSNDYWKRDNMFSLRYCNWNKDMFMGLLPNSQFGDVAV- 297
+DY +GS + + ++ D + N+F +RY NW +D+ G +P +Q+G+ +
Sbjct 222 IDYISGSADSL---QLDFTVEGFKDSF---NLFDMRYSNWQRDLLHGTIPQAQYGEASAV 275
Query 298 -VSG----VEGIDTFVPVEVFNSINETNIAKPPLTGTHTPVYTDDAMTSSTTPSRIRVAG 352
VSG VEG P + + +A L G T + + + T+ R+
Sbjct 276 PVSGSMQVVEG-----PTPPAFTTGQDGVAF--LNGNVTIQGSSGYLQAQTSVGESRILR 328
Query 353 GSGIPSSV---AGSALGVRSLLGGEFSILALRQAEALQKWKEITQSVDTNYRDQIKAHFG 409
+ S + S+ GV SILALR+AEA QKWKE+ + + +Y QI+AH+G
Sbjct 329 FNNTNSGLIVEGDSSFGV--------SILALRRAEAAQKWKEVALASEEDYPSQIEAHWG 380
Query 410 INTPASMSHMAQYIGGIARNLDISEVVNNNLSETGSEAV-IYGKGVGTGSGKMRYHTGSQ 468
+ + S M Q++G I +L I+EVVNNN+ TG A I GKG +G+G + ++ G Q
Sbjct 381 QSVNKAYSDMCQWLGSINIDLSINEVVNNNI--TGENAADIAGKGTMSGNGSINFNVGGQ 438
Query 469 YCIIMCIYHAVPLLDYAISGQDSQLLCTSVEDLPIPEFDNIGMEAVPAITLFNS-NAFDN 527
Y I+MC++H +P LDY S T+V D PIPEFD IGME VP I N D
Sbjct 439 YGIVMCVFHVLPQLDYITSAPHFGTTLTNVLDFPIPEFDKIGMEQVPVIRGLNPVKPKDG 498
Query 528 DLE-SDFDFLGYNPRYWPWKSKIDRVHGAFLTTLKDWVAPIDDFYL----NRWFASGGSS 582
D + S + GY P+Y+ WK+ +D+ G F +LK W+ P DD L + F +
Sbjct 499 DFKVSPNLYFGYAPQYYNWKTTLDKSMGEFRRSLKTWIIPFDDEALLAADSVDFPDNPNV 558
Query 583 QA-SISWPFFKVNPNTLDSIFAVAADSTWESDQLLINCDVSCKVVRPLSQDGMPY 636
+A S+ FFKV+P+ LD++FAV A+S +DQ L + VVR L +G+PY
Sbjct 559 EADSVKAGFFKVSPSVLDNLFAVKANSDLNTDQFLCSTLFDVNVVRSLDPNGLPY 613
>gi|575094321|emb|CDL65708.1| unnamed protein product [uncultured bacterium]
Length=642
Score = 284 bits (726), Expect = 1e-82, Method: Compositional matrix adjust.
Identities = 213/661 (32%), Positives = 328/661 (50%), Gaps = 51/661 (8%)
Query 2 AHFTGLKELQNHPHKAGFDIGSKNLFTAKVGELLPVYWDFAIPSCDYDIDLAYFTRTRPV 61
++ GL L+N P + FD+ +N+FTAKVGELLP + P + +YFTRT P+
Sbjct 5 SNIMGLHGLKNKPSRNSFDLSHRNMFTAKVGELLPCFVQELNPGDSVKVSSSYFTRTAPL 64
Query 62 QTAAYTRIREYFDFYAVPCDLLWKSFDSAVIQM------GEVAPVQAKTPLDPLTVGTDI 115
Q+ A+TR+RE ++ VP LWK FDS V+ M G+++ + A + + V T +
Sbjct 65 QSNAFTRLRENVQYFFVPYSALWKYFDSQVLNMTKNANGGDISRI-ASSLVGNQKVTTQM 123
Query 116 PWCTLSDLYTSLIFMNNRVSLGQTVSVVPDYSNIFGYSRCDTSHKLLLYLNYGNFVEPSS 175
P L+ L+ NR ++G SV P+++ G R S KLL L YGNF E +
Sbjct 124 PCVNYKTLHAYLLKFINRSTVGSDGSVGPEFNR--GCYRHAESAKLLQLLGYGNFPEQFA 181
Query 176 SNVGTSTNRWFNTSFTSSAVQNYSQ-KYSQNVSVSLFPLLAYQKIYQDFFRWSQWENADP 234
N N + + QN+ Y+ + +S+F LLAY KI D + + QW+ +
Sbjct 182 -------NFKVNNDKHNQSGQNFKDVTYNNSPYLSIFRLLAYHKICNDHYLYRQWQPYNA 234
Query 235 TAYNVDYYN-GSGNLFGNGGIASSIPSSNDYWKRDNMFSLRYCNWNKDMFMGLLPNSQFG 293
+ NVDY S +L SIP + ++ N+ +R+ N D F G+LP SQFG
Sbjct 235 SLCNVDYLTPNSSSLLSIDDALLSIPDDSIKAEKLNLLDMRFSNLPLDYFTGVLPTSQFG 294
Query 294 DVAVVSGVEGIDTFVPVEVFNSINETNIAKPPLTGTHTPVYTDDAMTSSTTPSRIRVAGG 353
+VV+ G + V +N T T T + + +S+ +++
Sbjct 295 SESVVNLNLGNASGSAV-----LNGTTSKDSGRWRTTTGEWEMEQRVASSANGNLKLDNS 349
Query 354 SGI----PSSVAGSALGVRSLLGGEFSILALRQAEALQKWKEITQSVDTNYRDQIKAHFG 409
+G + +G+ + + + L G SI+ALR A A QK+KEI + D +++ Q++AHFG
Sbjct 350 NGTFISHDHTFSGN-VAINTSLSGNLSIIALRNALAAQKYKEIQLANDVDFQSQVEAHFG 408
Query 410 INTPASMSHMAQYIGGIARNLDISEVVNNNLSETGSEAVIYGKG-VGTGSGKMRYHTGSQ 468
I P + + +IGG + ++I+E +N NLS G YG G GS +++ T
Sbjct 409 I-KPDEKNENSLFIGGSSSMININEQINQNLS--GDNKATYGAAPQGNGSASIKF-TAKT 464
Query 469 YCIIMCIYHAVPLLDYAISGQDSQLLCTSVEDLPIPEFDNIGME-------AVPAITLFN 521
Y +++ IY P+LD+A G D L T D IPE D+IGM+ A PA
Sbjct 465 YGVVIGIYRCTPVLDFAHLGIDRTLFKTDASDFVIPEMDSIGMQQTFRCEVAAPAPYNDE 524
Query 522 SNAF---DNDLESDFDFLGYNPRYWPWKSKIDRVHGAFLTTLKDWVAPI--DDFYLNRWF 576
AF D + GY PRY +K+ DR +GAF +LK WV I D N W
Sbjct 525 FKAFRVGDGSSPDMSETYGYAPRYSEFKTSYDRYNGAFCHSLKSWVTGINFDAIQNNVW- 583
Query 577 ASGGSSQASISWP-FFKVNPNTLDSIFAVAADSTWESDQLLINCDVSCKVVRPLSQDGMP 635
++ A I+ P F P+ + ++F V++ + + DQL + C R LS+ G+P
Sbjct 584 ----NTWAGINAPNMFACRPDIVKNLFLVSSTNNSDDDQLYVGMVNMCYATRNLSRYGLP 639
Query 636 Y 636
Y
Sbjct 640 Y 640
>gi|517172762|ref|WP_018361580.1| hypothetical protein [Prevotella nanceiensis]
Length=568
Score = 192 bits (488), Expect = 9e-50, Method: Compositional matrix adjust.
Identities = 172/649 (27%), Positives = 283/649 (44%), Gaps = 129/649 (20%)
Query 16 KAGFDIGSKNLFTAKVGELLPVYWDFAIPSCDYDIDLAYFTRTRPVQTAAYTRIREYFDF 75
+ FDI ++LFTA G LLPV +P +I+ + F RT P+ +AA+ +R ++F
Sbjct 18 RNAFDISQRHLFTAPAGALLPVLSLDLLPHDHVEINASDFMRTLPMNSAAFMSMRGVYEF 77
Query 76 YAVPCDLLWKSFDSAVIQMGE-----VAPVQAKTPLDPLTVGTDIP----WCTLSDLYTS 126
Y VP LW FD + M + + + KTP P V D+ WC +
Sbjct 78 YFVPYKQLWSGFDQFITGMSDYKSSFMYAFKGKTP--PSCVSFDVQKLVDWCKTNTA--- 132
Query 127 LIFMNNRVSLGQTVSVVPDYSNIFGYSRCDTSHKLLLYLNYGN--------FVEPSSSNV 178
+I G+ + +++L L YG + P+S+ +
Sbjct 133 --------------------KDIHGFDKNKGVYRILDLLGYGKYANSAGVPYTNPTSTTM 172
Query 179 GTSTNRWFNTSFTSSAVQNYSQKYSQNVSVSLFPLLAYQKIYQDFFRWSQWENADPTAYN 238
G T F LAYQKIY DF+R + +E ++N
Sbjct 173 GKCTP---------------------------FRGLAYQKIYNDFYRNTTYEEYQLESFN 205
Query 239 VDYYNGSGNLFGNGGIASSIPSSNDYWKRDNMFSLRYCNWNKDMFMGLLPNSQFGDVAVV 298
VD + +G+G + +IP N+ W D F+LRY N KD+ + P F
Sbjct 206 VDMF------YGSGKVKETIP--NEPWDYD-WFTLRYRNAQKDLLTNVRPTPLF------ 250
Query 299 SGVEGIDTFVPVEVFNSINETNIAKPPLTGTHTPVYTDDAMTSSTTPSRIRVAGGSGIPS 358
ID F P + F ++ + K P T Y D + +G+ S
Sbjct 251 ----SIDDFNP-QFFTGGSDIVMEKGPNVTGGTHEYRDSVVIVGKNLKE------NGVDS 299
Query 359 SVAGSALGVRSLLGGEFSILALRQAEALQKWKEITQSVDTNYRDQIKAHFGINTPASMSH 418
R+++ S+ +R A AL+K +T Y++Q++AHFGI+
Sbjct 300 K--------RTMI----SVADIRNAFALEKLASVTMRAGKTYKEQMEAHFGISVEEGRDG 347
Query 419 MAQYIGGIARNLDISEVVNNN-LSETGSEAVIY--------GKGVGTGSGKMRYHTGSQY 469
YIGG N+ + +V ++ + TG++ + GK G+GSG +R+ ++
Sbjct 348 RCTYIGGFDSNIQVGDVTQSSGTTVTGTKDTSFGGYLGRTTGKATGSGSGHIRFD-AKEH 406
Query 470 CIIMCIYHAVPLLDYAISGQDSQLLCTSVEDLPIPEFDNIGMEAVPAITL---FNSNAFD 526
I+MCIY VP + Y D + D +PEF+N+GM+ + A + +N+N +
Sbjct 407 GILMCIYSLVPDVQYDSKRVDPFVQKIERGDFFVPEFENLGMQPLFAKNISYKYNNNTAN 466
Query 527 NDLESDFDFLGYNPRYWPWKSKIDRVHGAFLTTLKDWVAPIDDFYLNRWFASGGSSQASI 586
+ ++ + G+ PRY +K+ +D HG F+ P+ + + R + G S ++
Sbjct 467 SRIK-NLGAFGWQPRYSEYKTALDINHGQFVHQ-----EPLSYWTVAR---ARGESMSNF 517
Query 587 SWPFFKVNPNTLDSIFAVAADSTWESDQLLINCDVSCKVVRPLSQDGMP 635
+ FK+NP LD +FAV + T +DQ+ C + V +S DGMP
Sbjct 518 NISTFKINPKWLDDVFAVNYNGTELTDQVFGGCYFNIVKVSDMSIDGMP 566
>gi|494308783|ref|WP_007173938.1| hypothetical protein [Prevotella bergensis]
gi|270333035|gb|EFA43821.1| putative capsid protein (F protein) [Prevotella bergensis DSM
17361]
Length=553
Score = 190 bits (483), Expect = 4e-49, Method: Compositional matrix adjust.
Identities = 171/641 (27%), Positives = 277/641 (43%), Gaps = 109/641 (17%)
Query 7 LKELQNHPHKAGFDIGSKNLFTAKVGELLPVYWDFAIPSCDYDIDLAYFTRTRPVQTAAY 66
+K + + ++ FD+ ++LFTA G LLPV IP +I+ F RT P+ TAA+
Sbjct 8 IKATRPNRNRNAFDLSQRHLFTAHAGMLLPVLNLDLIPHDHVEINAQDFMRTLPMNTAAF 67
Query 67 TRIREYFDFYAVPCDLLWKSFDSAVIQMGEVAPVQAKTPLDPLTVGTDIPWCTLSDLYTS 126
+R ++F+ VP LW FD + M + A + T +P+ + ++ S
Sbjct 68 ASMRGVYEFFFVPYHQLWAQFDQFITGMNDFHS-SANKSIQGGTSPLQVPYFNVDSVFNS 126
Query 127 LIFMNNRVSLGQTVSVVPDYSNIFGYSRCDTSHKLLLYLNYGNFVEPSSSNVGTSTNRWF 186
L N G + Y +G + +LL L YG R F
Sbjct 127 L---NTGKESGSGSTDDLQYKFKYG------AFRLLDLLGYG---------------RKF 162
Query 187 NTSFTSSAVQNYSQ-KYSQNVSVSLFPLLAYQKIYQDFFRWSQWENADPTAYNVDYYNGS 245
+ SF ++ N S K + + + S+F +LAY KIYQD++R S +EN D ++N D + G
Sbjct 163 D-SFGTAYPDNVSGLKNNLDYNCSVFRILAYNKIYQDYYRNSNYENFDTDSFNFDKFKG- 220
Query 246 GNLFGNGGIASSIPSSNDYWKRDNMFSLRYCNWNKDMFMGLLPNSQFGDVAVVSGVEGID 305
G + + + + ++F LRY N D F L SQ
Sbjct 221 ------GLVDAKVVA--------DLFKLRYRNAQTDYFTNLR-QSQL------------- 252
Query 306 TFVPVEVFNSINETNIAKPPLTGTHTPVYTDDAMTSSTTPSRIRVAGGSGIPSSVAGSAL 365
F F ++ NIA + +T RV G SS
Sbjct 253 -FSFTTAFEDVDNINIAPRDYVKSDGSNFT-------------RVNFGVDTDSS------ 292
Query 366 GVRSLLGGEFSILALRQAEALQKWKEITQSVDTNYRDQIKAHFGINTPASMSHMAQYIGG 425
G+FS+ +LR A A+ K +T ++DQ++AH+G+ P S Y+GG
Sbjct 293 ------EGDFSVSSLRAAFAVDKLLSVTMRAGKTFQDQMRAHYGVEIPDSRDGRVNYLGG 346
Query 426 IARNLDISEVVNNNLS-------ETGSEAVIYGKGVGTGSGKMRYHTGSQYCIIMCIYHA 478
++ +S+V + + E G + GKG G+G G++ + ++ ++MCIY
Sbjct 347 FDSDMQVSDVTQTSGTTATEYKPEAGYLGRVAGKGTGSGRGRIVFD-AKEHGVLMCIYSL 405
Query 479 VPLLDYAISGQDSQLLCTSVEDLPIPEFDNIGMEAVPA--ITLFNSNAFDNDLESDFDFL 536
VP + Y + D + D PEF+N+GM+ + + I+ F + N + L
Sbjct 406 VPQIQYDCTRLDPMVDKLDRFDYFTPEFENLGMQPLNSSYISSFCTTDPKNPV------L 459
Query 537 GYNPRYWPWKSKIDRVHGAFLTT--LKDWVAPIDDFYLNRWFASGGSSQASISWPFFKVN 594
GY PRY +K+ +D HG F + L W RW ++ + FK++
Sbjct 460 GYQPRYSEYKTALDVNHGQFAQSDALSSWSVS----RFRRW-----TTFPQLEIADFKID 510
Query 595 PNTLDSIFAVAADSTWESDQLLINCDVSCKVVRPLSQDGMP 635
P L+SIF V + T +D + C+ + V +S DGMP
Sbjct 511 PGCLNSIFPVDYNGTEANDCVYGGCNFNIVKVSDMSVDGMP 551
>gi|496521299|ref|WP_009229582.1| capsid protein [Prevotella sp. oral taxon 317]
gi|288330570|gb|EFC69154.1| putative capsid protein (F protein) [Prevotella sp. oral taxon
317 str. F0108]
Length=541
Score = 178 bits (451), Expect = 6e-45, Method: Compositional matrix adjust.
Identities = 168/642 (26%), Positives = 268/642 (42%), Gaps = 135/642 (21%)
Query 12 NHPHKAGFDIGSKNLFTAKVGELLPVYWDFAIPSCDYDIDLAYFTRTRPVQTAAYTRIRE 71
N P A FD+ K+L+TA G LLPV + I F RT P+ +AA+ +R
Sbjct 15 NRPRSA-FDLSQKHLYTAPAGALLPVLSVDLMFHDHIRIQAQDFMRTMPMNSAAFISMRG 73
Query 72 YFDFYAVPCDLLWKSFDSAVIQMGE-----VAPVQAKTPLDPLTVGTDIPWCTLSDLYTS 126
++F+ VP LW +D + M + V+ LD +P L+D+Y
Sbjct 74 VYEFFFVPYSQLWHPYDQFITSMNDYRSSVVSSAAGDKALD------SVPNVKLADMYK- 126
Query 127 LIFMNNRVSLGQTVSVVPDYSNIFGYSRCDTSHKLLLYLNYGNFVEPSSSNVGTSTNRWF 186
F+ R +IFGY + S +L+ L YG + S + V
Sbjct 127 --FVRERTD-----------KDIFGYPHSNNSCRLMDLLGYGKPITSSKTPV-------- 165
Query 187 NTSFTSSAVQNYSQKYSQNVSVSLFPLLAYQKIYQDFFRWSQWENADPTAYNVDYYNGSG 246
Y+ NV+ LF LLAY KIY D++R + +E D ++N+D+ G+
Sbjct 166 ------------PLLYTGNVN--LFRLLAYNKIYSDYYRNTTYEGVDVYSFNIDHKKGT- 210
Query 247 NLFGNGGIASSIPSSNDYWKRDNMFSLRYCNWNKDMFMGLLPNSQFGDVAVVSGVEGIDT 306
+P+++++ K N L Y N D + L P
Sbjct 211 ----------FVPTADEFKKYLN---LHYRNAPLDFYTNLRP------------------ 239
Query 307 FVPVEVFNSINETNIAKPPLTGTHTPVYTDDAMTSSTTPSRIRVAGGSGIPSSVAGSALG 366
TP++T + + S+ G +G S G++
Sbjct 240 ------------------------TPLFTIGSDSFSSVLQLSDPTGSAGF--SADGNSAK 273
Query 367 VRSLLGGEFSILALRQAEALQKWKEITQSVDTNYRDQIKAHFGINTPASMSHMAQYIGGI 426
+ ++ A+R A AL K I+ Y +QI+AHFG+ Y+GG
Sbjct 274 LNMASPDVLNVSAIRSAFALDKLLSISMRAGKTYAEQIEAHFGVTVSEGRDGQVYYLGGF 333
Query 427 ARNLDISEV------VNNNLSETGSEAV------IYGKGVGTGSGKMRYHTGSQYCIIMC 474
N+ + +V N N+SE G+ + I GKG G+G G++++ + ++MC
Sbjct 334 DSNVQVGDVTQTSGTTNPNVSEVGNAKLAGYLGKITGKGTGSGYGEIQFD-AKEPGVLMC 392
Query 475 IYHAVPLLDYAISGQDSQLLCTSVEDLPIPEFDNIGMEA-VPAITLFNSNAFDNDLESDF 533
IY VP + Y D + + D IPEF+N+GM+ VPA N A DN
Sbjct 393 IYSVVPAMQYDCMRLDPFVAKQTRGDYFIPEFENLGMQPIVPAFVSLN-RAKDNS----- 446
Query 534 DFLGYNPRYWPWKSKIDRVHGAFLTTLKDWVAPIDDFYLNRWFASGGSSQASISWPFFKV 593
G+ PRY +K+ D HG F P+ + + R A G + + + K+
Sbjct 447 --YGWQPRYSEYKTAFDINHGQFANG-----EPLSYWSIAR--ARGSDTLNTFNVAALKI 497
Query 594 NPNTLDSIFAVAADSTWESDQLLINCDVSCKVVRPLSQDGMP 635
NP+ LDS+FAV + T +D + + + V +++DGMP
Sbjct 498 NPHWLDSVFAVNYNGTEVTDCMFGYAHFNIEKVSDMTEDGMP 539
>gi|494306153|ref|WP_007173049.1| hypothetical protein [Prevotella bergensis]
gi|270333881|gb|EFA44667.1| putative capsid protein (F protein) [Prevotella bergensis DSM
17361]
Length=519
Score = 177 bits (449), Expect = 8e-45, Method: Compositional matrix adjust.
Identities = 161/614 (26%), Positives = 262/614 (43%), Gaps = 110/614 (18%)
Query 34 LLPVYWDFAIPSCDYDIDLAYFTRTRPVQTAAYTRIREYFDFYAVPCDLLWKSFDSAVIQ 93
LLPV IP +I+ F RT P+ TAA+ +R ++F+ VP LW FD +
Sbjct 2 LLPVLNLDLIPHDHVEINAQDFMRTLPMNTAAFASMRGVYEFFFVPYHQLWAQFDQFITG 61
Query 94 MGEVAPVQAKTPLDPLTVGTDIPWCTLSDLYTSLIFMNNRVSLGQTVSVVPDYSNIFGYS 153
M + K+ + T +P+ L ++ ++I T S D F Y
Sbjct 62 MNDFHSSANKS-IQGGTSPLQVPYFNLESVFKNII------ERDSTPSFQDDLQYRFKYG 114
Query 154 RCDTSHKLLLYLNYGNFVEPSSSNVGTSTNRWFNTSFTSSAVQNYSQ-KYSQNVSVSLFP 212
+ +LL L YG R F+ SF ++ N S K + + + S+F
Sbjct 115 ----AFRLLDLLGYG---------------RKFD-SFGTAYPDNVSGLKNNLDYNCSVFR 154
Query 213 LLAYQKIYQDFFRWSQWENADPTAYNVDYYNGSGNLFGNGGIASSIPSSNDYWKRDNMFS 272
+LAY KIYQD++R S +EN D ++N D + G G + + + + ++F
Sbjct 155 VLAYNKIYQDYYRNSNYENFDTDSFNFDKFKG-------GLVDAKVVA--------DLFK 199
Query 273 LRYCNWNKDMFMGLLPNSQFGDVAVVSGVEGIDTFVPVEVFNSINETNIAKPPLTGTHTP 332
LRY N D F L + F + S E ++ F+ + +K T + P
Sbjct 200 LRYRNAQTDYFTNLRQSQLFTFIPEFSDDEHLN-------FDRDQYADQSKSNFTQLNFP 252
Query 333 VYTDDAMTSSTTPSRIRVAGGSGIPSSVAGSALGVRSLLGGEFSILALRQAEALQKWKEI 392
V D+ + G FS+ +LR A A+ K +
Sbjct 253 VDVDNNL---------------------------------GYFSVSSLRSAFAVDKLLSV 279
Query 393 TQSVDTNYRDQIKAHFGINTPASMSHMAQYIGGIARNLDISEVVNNNLS-------ETGS 445
T ++DQ++AH+G+ P S Y+GG +L +S+V + + E G
Sbjct 280 TMRAGKTFQDQMRAHYGVEIPDSRDGRVNYLGGFDSDLQVSDVTQTSGTTATEYKPEAGY 339
Query 446 EAVIYGKGVGTGSGKMRYHTGSQYCIIMCIYHAVPLLDYAISGQDSQLLCTSVEDLPIPE 505
I GKG G+G G++ + ++ ++MCIY VP + Y + D + D PE
Sbjct 340 LGRIAGKGTGSGRGRIVFD-AKEHGVLMCIYSLVPQIQYDCTRLDPMVDKLDRFDFFTPE 398
Query 506 FDNIGMEAVPA--ITLFNSNAFDNDLESDFDFLGYNPRYWPWKSKIDRVHGAFLT--TLK 561
F+N+GM+ + + I+ F + N + LGY PRY +K+ +D HG F L
Sbjct 399 FENLGMQPLNSSYISSFCTPDPKNPV------LGYQPRYSEYKTALDINHGQFAQNDALS 452
Query 562 DWVAPIDDFYLNRWFASGGSSQASISWPFFKVNPNTLDSIFAVAADSTWESDQLLINCDV 621
W RW ++ + FK++P L+S+F V + T +D + C+
Sbjct 453 SWSVS----RFRRW-----TTFPQLEIADFKIDPGCLNSVFPVEFNGTESTDCVFGGCNF 503
Query 622 SCKVVRPLSQDGMP 635
+ V +S DGMP
Sbjct 504 NIVKVSDMSVDGMP 517
Lambda K H a alpha
0.319 0.135 0.419 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 4793916891504