bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: All non-redundant GenBank CDS translations+PDB+SwissProt+PIR+PRF
excluding environmental samples from WGS projects
49,011,213 sequences; 17,563,301,199 total letters
Query= Contig-30_CDS_annotation_glimmer3.pl_2_2
Length=555
Score E
Sequences producing significant alignments: (Bits) Value
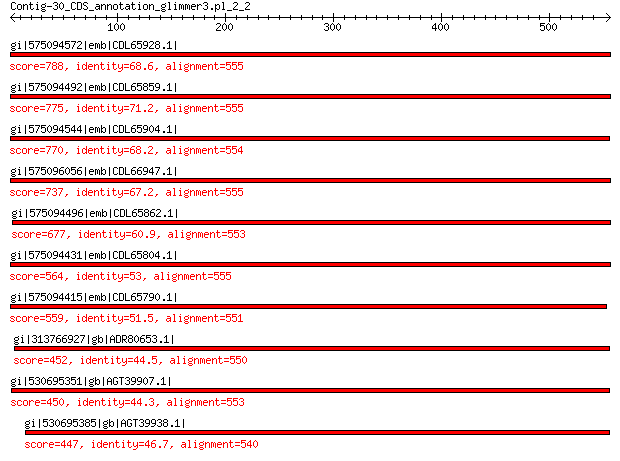
gi|575094572|emb|CDL65928.1| unnamed protein product 788 0.0
gi|575094492|emb|CDL65859.1| unnamed protein product 775 0.0
gi|575094544|emb|CDL65904.1| unnamed protein product 770 0.0
gi|575096056|emb|CDL66947.1| unnamed protein product 737 0.0
gi|575094496|emb|CDL65862.1| unnamed protein product 677 0.0
gi|575094431|emb|CDL65804.1| unnamed protein product 564 0.0
gi|575094415|emb|CDL65790.1| unnamed protein product 559 0.0
gi|313766927|gb|ADR80653.1| putative major coat protein 452 2e-149
gi|530695351|gb|AGT39907.1| major capsid protein 450 1e-148
gi|530695385|gb|AGT39938.1| major capsid protein 447 5e-148
>gi|575094572|emb|CDL65928.1| unnamed protein product [uncultured bacterium]
Length=556
Score = 788 bits (2036), Expect = 0.0, Method: Compositional matrix adjust.
Identities = 381/559 (68%), Positives = 450/559 (81%), Gaps = 7/559 (1%)
Query 1 MNRNTNSHFALNPTRIDMSRSTFDRNSSVKTSFNVGDIVPFYVDEVLPGDTFDIDTSKVI 60
MNRN SHFA NPT ID+SRSTFDR+SSVK +FN G+I+PF+++EVLPGDTF + TSKVI
Sbjct 1 MNRNVESHFAKNPTNIDISRSTFDRSSSVKLTFNTGEIIPFFIEEVLPGDTFKVKTSKVI 60
Query 61 RMPSLLTPIMDNLYLDTYYFFVPNRIVWQHWKELMGENNESAWIPTTEYEVPQITAPSGG 120
R+ +LLTP+MDN+YLDTYYFFVPNR+VW+HWKE GEN +SAWIP EY++PQ+TAP GG
Sbjct 61 RLQTLLTPMMDNIYLDTYYFFVPNRLVWEHWKEFNGENTQSAWIPEVEYQIPQLTAPEGG 120
Query 121 WSIGTIADYMGVPTGVSGLSVNALPFRAYALICNEWFRDENLCDPLNIPLTDATVAGVNT 180
W+IGT+ADY G+PTGVSG+SVNALPFRAYAL+CNEWFRD+NL DPLNIP+ DATV GVNT
Sbjct 121 WNIGTLADYFGIPTGVSGISVNALPFRAYALVCNEWFRDQNLSDPLNIPVGDATVTGVNT 180
Query 181 GTFVTDVAKGGLPYKAAKYRDYFTSCLPAPQKSEDVTIPVSSGANYPVLSLS---DIVP- 236
GTF+TDV KGGLPY AAKY DYFTSCLPAPQK DVTIPV+SG N PV+ L+ D P
Sbjct 181 GTFITDVVKGGLPYTAAKYHDYFTSCLPAPQKGPDVTIPVTSGHNLPVMFLNETHDAGPY 240
Query 237 TPGTVPVKWNDANNVVSDAQWLLGGKNYNGTITSNDISLTKTNTGPTYSAVTPINLWAVN 296
P V ++ ++ N G + + T ++ ++ T G + TP N+WAV
Sbjct 241 KPFGVGIQNSELRNFYGFGSGSSGATSTSDTSSTVEVGSDGTGIGQNF--WTPTNMWAVE 298
Query 297 DGSVSSATINQLRLAFQVQKLYERDARGGTRYIEVLKSHFGVTSPDARLQRPEYLGGNRI 356
G V ATINQLRLAFQ+QKLYE+DARGGTRY E+++SHFGV SPD+RLQRPEYLGGNRI
Sbjct 299 SGDVGMATINQLRLAFQLQKLYEKDARGGTRYTEIIRSHFGVVSPDSRLQRPEYLGGNRI 358
Query 357 PIVISEINQTSGTSANSTPQGNPSGQSRTTDVHSDFKKSFVEHGFIIGVMVARYDHTYQQ 416
PI +++I Q S S +P G +G S TTD +SDF KSFVEHG+IIG++VARYDHTYQQ
Sbjct 359 PINVNQIIQQS-QSTEQSPLGALAGMSVTTDKNSDFIKSFVEHGYIIGLVVARYDHTYQQ 417
Query 417 GLERFWSRKGRLDYYWPVFANIGEQAVLNKEIYAQGNGTDDEVFGYQEAWADYRYKPNRV 476
GL+R WSRK R D+YWPV ANIGEQAVLNKEIY G+ TDDEVFGYQEAWA+YRYKPNRV
Sbjct 418 GLDRMWSRKDRFDFYWPVLANIGEQAVLNKEIYIDGSDTDDEVFGYQEAWAEYRYKPNRV 477
Query 477 TGEMRSQAPQSLDVWHLGDDYSKLPSLSDSWVQEDSAVVNRVIAVSEENSNQLWADIFIK 536
GEMRS APQSLDVWHLGDDYS LP LSDSW++ED V+RV+AV+ S+QL+ADI+I
Sbjct 478 CGEMRSSAPQSLDVWHLGDDYSSLPYLSDSWIREDKTNVDRVLAVTSSVSDQLFADIYIC 537
Query 537 NKCTRAMPMYSIPGLIDHH 555
NK TR MPMYSIPGLIDHH
Sbjct 538 NKATRPMPMYSIPGLIDHH 556
>gi|575094492|emb|CDL65859.1| unnamed protein product [uncultured bacterium]
Length=551
Score = 775 bits (2001), Expect = 0.0, Method: Compositional matrix adjust.
Identities = 395/567 (70%), Positives = 451/567 (80%), Gaps = 28/567 (5%)
Query 1 MNRNTNSHFALNPTRIDMSRSTFDRNSSVKTSFNVGDIVPFYVDEVLPGDTFDIDTSKVI 60
M RNTNS FALNPTR+DMSRS FDR+SS KT+FNVGD++PFYVDE+LPGDTF IDTSKV+
Sbjct 1 MTRNTNSRFALNPTRLDMSRSRFDRSSSYKTTFNVGDLIPFYVDEILPGDTFSIDTSKVV 60
Query 61 RMPSLLTPIMDNLYLDTYYFFVPNRIVWQHWKELMGENNESAWIPTTEYEVPQITAPSGG 120
RM SLLTP+MDN+YLDTY+FFVPNR+ W HW+ELMGEN +SAW P EY VPQITAP GG
Sbjct 61 RMQSLLTPVMDNIYLDTYFFFVPNRLTWSHWRELMGENTQSAWTPQVEYSVPQITAPEGG 120
Query 121 WSIGTIADYMGVPTGVSGLSVNALPFRAYALICNEWFRDENLCDPLNIPLTDATVAGVNT 180
W++GTIADYMG+PTGVSGLSVNA+PFRAYALICNEWFRDENL DPLNIP+ DATVAGVNT
Sbjct 121 WNVGTIADYMGIPTGVSGLSVNAMPFRAYALICNEWFRDENLTDPLNIPVGDATVAGVNT 180
Query 181 GTFVTDVAKGGLPYKAAKYRDYFTSCLPAPQKSEDVTI-PVSSGANYPVLSLSDIVPTPG 239
GT+VTDVAKGGLP+KAAKY DYFTSCLPAPQK DV I V SG IVP
Sbjct 181 GTYVTDVAKGGLPFKAAKYHDYFTSCLPAPQKGPDVLISAVGSG----------IVPVTA 230
Query 240 TVPVKWNDANNVVSDAQWLLGGK----NYNGTITSNDISLTKT--NTGPTYS-AVTPINL 292
T ND+ NV S +G NY + +T T + P + ++ P NL
Sbjct 231 T--DNDNDSLNVNSPGMRFVGNSSTSVNYLAFGGGDGYVVTDTPKPSTPIHGISMIPTNL 288
Query 293 WAVNDGSVSS----ATINQLRLAFQVQKLYERDARGGTRYIEVLKSHFGVTSPDARLQRP 348
WA D S ++ ATINQLR AFQ+QKLYERDARGGTRYIE+LKSHFGVTSPDARLQRP
Sbjct 289 WA--DLSTATDLPVATINQLRTAFQIQKLYERDARGGTRYIEILKSHFGVTSPDARLQRP 346
Query 349 EYLGGNRIPIVISEINQTSGTSANSTPQGNPSGQSRTTDVHSDFKKSFVEHGFIIGVMVA 408
EYLGG+R+PI I+++ Q+S T A TPQGN + S TTD HS+F KSFVEHGFIIG+MVA
Sbjct 347 EYLGGSRVPININQVIQSSETGA--TPQGNAAAYSLTTDSHSEFTKSFVEHGFIIGLMVA 404
Query 409 RYDHTYQQGLERFWSRKGRLDYYWPVFANIGEQAVLNKEIYAQGNGTDDEVFGYQEAWAD 468
RYDH+YQQGL+RFWSRK R DYYWPVFAN+GE AV NKEI+AQG DDEVFGYQEAWAD
Sbjct 405 RYDHSYQQGLQRFWSRKDRFDYYWPVFANLGEMAVKNKEIFAQGTDVDDEVFGYQEAWAD 464
Query 469 YRYKPNRVTGEMRSQAPQSLDVWHLGDDYSKLPSLSDSWVQEDSAVVNRVIAVSEENSNQ 528
YRYKP+ VTGEMRSQ QSLD+WHL DDY LPSLSDSW++EDS+ VNRV+AVS+ S Q
Sbjct 465 YRYKPSVVTGEMRSQYAQSLDIWHLADDYENLPSLSDSWIREDSSTVNRVLAVSDSVSAQ 524
Query 529 LWADIFIKNKCTRAMPMYSIPGLIDHH 555
L+ DI+I+ TR MP+YSIPGLIDHH
Sbjct 525 LFCDIYIRCLATRPMPLYSIPGLIDHH 551
>gi|575094544|emb|CDL65904.1| unnamed protein product [uncultured bacterium]
Length=551
Score = 770 bits (1988), Expect = 0.0, Method: Compositional matrix adjust.
Identities = 378/566 (67%), Positives = 453/566 (80%), Gaps = 28/566 (5%)
Query 1 MNRNTNSHFALNPTRIDMSRSTFDRNSSVKTSFNVGDIVPFYVDEVLPGDTFDIDTSKVI 60
MNRN SHF+ P+ +D+SRS FDR+SS+KT+FNVGD++PFY+DEVLPGDTF++ +SKVI
Sbjct 1 MNRNVESHFSRLPS-VDISRSQFDRSSSLKTTFNVGDLIPFYIDEVLPGDTFNVKSSKVI 59
Query 61 RMPSLLTPIMDNLYLDTYYFFVPNRIVWQHWKELMGENNESAWIPTTEYEVPQITAPSGG 120
RM SL+TPIMDN+YLDTYYFFVPNR+VW HW++ GEN ESAW+PTTEY+VPQ+TAP+ G
Sbjct 60 RMQSLVTPIMDNIYLDTYYFFVPNRLVWSHWQQFNGENTESAWLPTTEYQVPQVTAPANG 119
Query 121 WSIGTIADYMGVPTGVSGLSVNALPFRAYALICNEWFRDENLCDPLNIPLTDATVAGVNT 180
WSIGTIADY G+PTGV+ SVNALPFRAYALICNEWFRDENL DPLNIP++DATV G N
Sbjct 120 WSIGTIADYFGIPTGVA-CSVNALPFRAYALICNEWFRDENLSDPLNIPISDATVVGSNG 178
Query 181 GTFVTDVAKGGLPYKAAKYRDYFTSCLPAPQKSEDVTIPVSSGANYPV-LSLSDIVPTP- 238
++TD+ KGG+P+KA KY DYFTSCLPAPQK DV +P+SS PV ++ SD + P
Sbjct 179 DNYITDIVKGGMPFKACKYHDYFTSCLPAPQKGPDVLLPLSSS---PVPVTTSDTMVDPL 235
Query 239 --GTVPV----KWNDA----NNVVSDAQWLLGGKNYNGTITSNDISLTKTNTGPTYSAVT 288
P+ WN + N++ + + G NY + DI PT A
Sbjct 236 QYSKYPMAGVDSWNLSPTLMRNIIRPFEGVEGA-NYQVHQFTGDI--------PTIDAFR 286
Query 289 PINLWAVNDGSVSSATINQLRLAFQVQKLYERDARGGTRYIEVLKSHFGVTSPDARLQRP 348
P+NL A N + ++A+INQLRLAFQ+Q+LYERDARGGTRYIE+LKSHFGVTSPDARLQRP
Sbjct 287 PLNLVA-NLQNATAASINQLRLAFQIQRLYERDARGGTRYIEILKSHFGVTSPDARLQRP 345
Query 349 EYLGGNRIPIVISEINQTSGTSANSTPQGNPSGQSRTTDVHSDFKKSFVEHGFIIGVMVA 408
EYLGGNRIPI I+++ Q S T++ S PQGNP GQS TTD ++DF KSFVEHGF+IG+MVA
Sbjct 346 EYLGGNRIPININQVLQQSETTSTS-PQGNPVGQSLTTDTNADFVKSFVEHGFVIGLMVA 404
Query 409 RYDHTYQQGLERFWSRKGRLDYYWPVFANIGEQAVLNKEIYAQGNGTDDEVFGYQEAWAD 468
RYDHTYQQGLERFWSRK R DYYWPVFA+IGEQAVLNKEIY G DDEVFGYQEA+AD
Sbjct 405 RYDHTYQQGLERFWSRKDRFDYYWPVFAHIGEQAVLNKEIYTSGTAVDDEVFGYQEAYAD 464
Query 469 YRYKPNRVTGEMRSQAPQSLDVWHLGDDYSKLPSLSDSWVQEDSAVVNRVIAVSEENSNQ 528
YRYKP+RVTGEMRS APQSLDVWHL DDY+ LPSLSDSW++E ++ V+RV+AVS S Q
Sbjct 465 YRYKPSRVTGEMRSAAPQSLDVWHLADDYASLPSLSDSWIRESASTVDRVLAVSSNVSAQ 524
Query 529 LWADIFIKNKCTRAMPMYSIPGLIDH 554
L+ DI+I+N+ TR MPMYS+PGLIDH
Sbjct 525 LFCDIYIQNRSTRPMPMYSVPGLIDH 550
>gi|575096056|emb|CDL66947.1| unnamed protein product [uncultured bacterium]
Length=570
Score = 737 bits (1903), Expect = 0.0, Method: Compositional matrix adjust.
Identities = 373/585 (64%), Positives = 443/585 (76%), Gaps = 46/585 (8%)
Query 1 MNRNTNSHFALNPTRIDMSRSTFDRNSSVKTSFNVGDIVPFYVDEVLPGDTFDIDTSKVI 60
MNRNT SHF+L P +D+SRS FDR+SS+KT+FN GD+VPF+++EVLPGDTF +D+SKV+
Sbjct 2 MNRNTESHFSLLP-HVDISRSRFDRSSSIKTTFNAGDVVPFFLEEVLPGDTFSVDSSKVV 60
Query 61 RMPSLLTPIMDNLYLDTYYFFVPNRIVWQHWKELMGENNESAWIPTTEYEVPQITAPSGG 120
RM +LLTP+MDN+YLDTYYFFVPNR+VWQHWKE GENNESAWIP TEY +PQ+ +P GG
Sbjct 61 RMQTLLTPMMDNVYLDTYYFFVPNRLVWQHWKEFCGENNESAWIPQTEYAIPQLKSPVGG 120
Query 121 WSIGTIADYMGVPTGVSGLSVNALPFRAYALICNEWFRDENLCDPLNIPLTDATVAGVNT 180
+ +GTIADY G+PTGV+ LSV+ALPFRAYALI NEWFRDENL DPL +P DATV GVNT
Sbjct 121 FEVGTIADYFGLPTGVANLSVSALPFRAYALIMNEWFRDENLMDPLVVPTDDATVTGVNT 180
Query 181 GTFVTDVAKGGLPYKAAKYRDYFTSCLPAPQKSEDVTIPVSSGANYPV------LSLSDI 234
G FVTDVAKGG P+ AAKY DYFTS LPAPQK DV IPV+S NY V L+LSD
Sbjct 181 GIFVTDVAKGGKPFVAAKYHDYFTSALPAPQKGPDVVIPVASAGNYNVVGNGKGLALSD- 239
Query 235 VPTPGTVPVKWNDANNVVSDAQWLLGGKNYNGT-----------------------ITSN 271
+++ + L G N GT + +
Sbjct 240 -----------GSKMSIICNG---LSGSNGQGTELFASGILGSQVGSSGGFGSGSSLRGD 285
Query 272 DISLTKTNTGPTYSAVTPINLWAVNDGSVSSATINQLRLAFQVQKLYERDARGGTRYIEV 331
I L + + L A+ G+ ++ATINQLR+AFQ+QK YE+ ARGG+RY EV
Sbjct 286 GIILGVPTAAQLGNNLENSGLIAIASGNAAAATINQLRMAFQIQKFYEKQARGGSRYTEV 345
Query 332 LKSHFGVTSPDARLQRPEYLGGNRIPIVISEINQTSGT-SANSTPQGNPSGQSRTTDVHS 390
++S FGVTSPDARLQR EYLGGNRIPI I+++ Q SGT SA++TPQG G S+TTD HS
Sbjct 346 IRSFFGVTSPDARLQRSEYLGGNRIPININQVIQQSGTGSASTTPQGTVVGMSQTTDTHS 405
Query 391 DFKKSFVEHGFIIGVMVARYDHTYQQGLERFWSRKGRLDYYWPVFANIGEQAVLNKEIYA 450
DF KSF EHGFIIGVM ARYDHTYQQG++R WSRK + DYYWPVF+NIGEQA+ NKEIYA
Sbjct 406 DFTKSFTEHGFIIGVMCARYDHTYQQGIDRMWSRKDKFDYYWPVFSNIGEQAIKNKEIYA 465
Query 451 QGNGTDDEVFGYQEAWADYRYKPNRVTGEMRSQAPQSLDVWHLGDDYSKLPSLSDSWVQE 510
QGN TDDEVFGYQEAWA+YRYKP+RVTGEMRS QSLDVWHL DDYSKLPSLSD W++E
Sbjct 466 QGNATDDEVFGYQEAWAEYRYKPSRVTGEMRSSYAQSLDVWHLADDYSKLPSLSDEWIRE 525
Query 511 DSAVVNRVIAVSEENSNQLWADIFIKNKCTRAMPMYSIPGLIDHH 555
D+ +NRV+AVS++NSNQ +ADI++KN CTR MPMYSIPGLIDHH
Sbjct 526 DAKTLNRVLAVSDQNSNQFFADIYVKNLCTRPMPMYSIPGLIDHH 570
>gi|575094496|emb|CDL65862.1| unnamed protein product [uncultured bacterium]
Length=568
Score = 677 bits (1748), Expect = 0.0, Method: Compositional matrix adjust.
Identities = 337/573 (59%), Positives = 420/573 (73%), Gaps = 26/573 (5%)
Query 3 RNTNSHFALNPTRIDMSRSTFDRNSSVKTSFNVGDIVPFYVDEVLPGDTFDIDTSKVIRM 62
RN NS F+ NP +D+ RSTF+R+S+ KTS N+G+++PFY DEVLPGDTF + T+KV+R+
Sbjct 2 RNENSRFSENPVTLDIQRSTFNRSSTYKTSANIGELIPFYYDEVLPGDTFQVKTNKVVRL 61
Query 63 PSLLTPIMDNLYLDTYYFFVPNRIVWQHWKELMGENNESAWIPTTEYEVPQITAP-SGGW 121
L++ MDNLY DTYYFFVPNR+VW+HW+E MGEN + AWIP TEY +PQIT+P S G+
Sbjct 62 QPLVSAPMDNLYFDTYYFFVPNRLVWEHWEEFMGENKQGAWIPQTEYTIPQITSPASTGF 121
Query 122 SIGTIADYMGVPTGVSGLSVNALPFRAYALICNEWFRDENLCDPLNIPLTDATVAGVNTG 181
IGTIADY G+PTGV LSV+ALPFRAYALI +EWFRD+NL PLNIPL D T+ GVNTG
Sbjct 122 EIGTIADYFGIPTGVPNLSVSALPFRAYALIVDEWFRDQNLQLPLNIPLDDTTLQGVNTG 181
Query 182 TFVTDVAKGGLPYKAAKYRDYFTSCLPAPQKSEDVTI------PVSSG----ANYPVLSL 231
+VTD KGG P+ AAKY DYFTSCLP+PQK DVTI PV +G N +L
Sbjct 182 DYVTDTVKGGKPFVAAKYHDYFTSCLPSPQKGPDVTIAAVGDFPVYTGDPHNNNGSNKAL 241
Query 232 SDIVPTPGTVPVKWNDANNVVSDAQWLLGGKNYN----GTITSNDISLTKTNTGPTYS-- 285
+ + V ++ N ++ L G + G + +++I++T + P S
Sbjct 242 HYGISNISSGSVSFSQGNYIIPSV--LTTGSTQSVPAQGKLNASNITMTTSPGSPDSSFG 299
Query 286 ---AVTPINLWAVNDGSVSSATINQLRLAFQVQKLYERDARGGTRYIEVLKSHFGVTSPD 342
+V P NL+A S ++ TINQLR+AFQ+QKLYE+DAR G+RY E+++SHF VT D
Sbjct 300 SKLSVYPDNLYA---SSGTATTINQLRMAFQIQKLYEKDARAGSRYRELIRSHFSVTPLD 356
Query 343 ARLQRPEYLGGNRIPIVISEINQTSGTSANSTPQGNPSGQSRTTDVHSDFKKSFVEHGFI 402
AR+Q PEYLGGNRIPI I+++ QTS TS + +PQGN +GQS T+D H DF KSF EHG +
Sbjct 357 ARMQVPEYLGGNRIPININQVVQTSQTS-DVSPQGNVAGQSLTSDSHGDFIKSFTEHGML 415
Query 403 IGVMVARYDHTYQQGLERFWSRKGRLDYYWPVFANIGEQAVLNKEIYAQGNGTDDEVFGY 462
IGV VARYDHTYQQG+ + WSRK R DYYWPV ANIGEQAVLNKEIYAQG D+EVFGY
Sbjct 416 IGVAVARYDHTYQQGVSKLWSRKTRFDYYWPVLANIGEQAVLNKEIYAQGTAQDEEVFGY 475
Query 463 QEAWADYRYKPNRVTGEMRSQAPQSLDVWHLGDDYSKLPSLSDSWVQEDSAVVNRVIAVS 522
QEAWA+YRYKP+ VTGEMRS A SLD WH DDY+ LP LS W++ED ++RV+AVS
Sbjct 476 QEAWAEYRYKPSIVTGEMRSSARTSLDSWHFADDYNSLPKLSADWIKEDKTNIDRVLAVS 535
Query 523 EENSNQLWADIFIKNKCTRAMPMYSIPGLIDHH 555
SNQ +AD +I+N+ TRA+P YSIPGLIDHH
Sbjct 536 SSVSNQYFADFYIENETTRALPFYSIPGLIDHH 568
>gi|575094431|emb|CDL65804.1| unnamed protein product [uncultured bacterium]
Length=560
Score = 564 bits (1454), Expect = 0.0, Method: Compositional matrix adjust.
Identities = 294/571 (51%), Positives = 375/571 (66%), Gaps = 27/571 (5%)
Query 1 MNRNTNSHFALNPTRIDMSRSTFDRNSSVKTSFNVGDIVPFYVDEVLPGDTFDIDTSKVI 60
MNRN+N +FA NP + +SRS F+R S +F+ G+IVP YVDEVLPGDTF++D + +I
Sbjct 1 MNRNSNFNFARNPG-VSLSRSRFNRTSDRLDTFDTGEIVPIYVDEVLPGDTFELDMTAII 59
Query 61 RMPSLLTPIMDNLYLDTYYFFVPNRIVWQHWKELMGENNESAWIPTTEYEVPQITAPSGG 120
R + + P+MDN +LD Y+FFVPNR+ W+HW+ELMGEN +AW +Y VPQ+TAP+GG
Sbjct 60 RGSTPIFPVMDNSFLDVYFFFVPNRLTWEHWRELMGENRTTAWTQPVDYSVPQVTAPAGG 119
Query 121 WSIGTIADYMGVPTGVSGLSVNALPFRAYALICNEWFRDENLCDPLNIPLTDATVAGVNT 180
W ++AD+MG+PT V +SVNALPFRAY LI NE+FR++NL +P + +TDA +AG N
Sbjct 120 WEELSLADHMGIPTKVDNISVNALPFRAYGLIYNEFFRNQNLTNPTQVEVTDANIAGKNP 179
Query 181 GTFVTD---VAKGGLPYKAAKYRDYFTSCLPAPQKSEDVTI-------PVSSGANYPVLS 230
G K+AK+ DYFT LP PQK E V I PV G + L
Sbjct 180 NDVKNSNDWAITGAKCLKSAKFFDYFTGALPQPQKGEPVEINLASSWLPVGIGDYHGPL- 238
Query 231 LSDIVPTPGTVPVKWNDANNVVSDAQ-WLLGGKNYNGTITSNDISLTKTNTGPTYS---- 285
D V T + W ++ + + + LG G + N + +T G ++S
Sbjct 239 --DKVSNSDT--LTWESPSSEGNTKRTYALGMVQQEGEVNPNGLKNFETKAGGSFSESGA 294
Query 286 -AVTPINLWAVNDGSVSSATINQLRLAFQVQKLYERDARGGTRYIEVLKSHFGVTSPDAR 344
A P NLWA ++AT+NQLR AFQVQKL E+DARGGTRY E+LK+HFGVT+ DAR
Sbjct 295 VAAYPTNLWA--SPVTAAATVNQLRQAFQVQKLLEKDARGGTRYREILKNHFGVTTSDAR 352
Query 345 LQRPEYLGGNRIPIVISEINQTSGTSANSTPQGNPSGQSRTTDVHSDFKKSFVEHGFIIG 404
+Q PEYLGG ++PI +S++ QTS S +++PQGN + S T S F KSF EHGFIIG
Sbjct 353 MQIPEYLGGCKVPINVSQVVQTSA-STDASPQGNTAAISVTPFSKSMFTKSFDEHGFIIG 411
Query 405 VMVARYDHTYQQGLERFWSRKGRLDYYWPVFANIGEQAVLNKEIYAQGNGTDDEVFGYQE 464
V AR +YQQG+ER WSRK RLDYY+PV ANIGEQA+LNKEIYAQGN DDE FGYQE
Sbjct 412 VATARTAQSYQQGIERMWSRKDRLDYYFPVLANIGEQAILNKEIYAQGNAKDDEAFGYQE 471
Query 465 AWADYRYKPNRVTGEMRSQAPQSLDVWHLGDDYSKLPSLSDSWVQEDSAVVNRVIAVSEE 524
AWADYRYKPN + G RS A QSLD WH G DY KLP+LS W+++ + R +AV E
Sbjct 472 AWADYRYKPNTICGRFRSNAQQSLDAWHYGQDYDKLPTLSTDWMEQSDIEMKRTLAVQTE 531
Query 525 NSNQLWADIFIKNKCTRAMPMYSIPGLIDHH 555
A+ K R MP+YSIPGLIDH+
Sbjct 532 PD--FIANFRFNCKTVRVMPLYSIPGLIDHN 560
>gi|575094415|emb|CDL65790.1| unnamed protein product [uncultured bacterium]
Length=569
Score = 559 bits (1440), Expect = 0.0, Method: Compositional matrix adjust.
Identities = 284/572 (50%), Positives = 363/572 (63%), Gaps = 27/572 (5%)
Query 1 MNRNTNSHFALNPTRIDMSRSTFDRNSSVKTSFNVGDIVPFYVDEVLPGDTFDIDTSKVI 60
MNRN +H++ P ++ R+ F R+ S T+ N GD+VP YVDEVLPGDT I ++
Sbjct 1 MNRNAEAHYSQIP-HANIQRAKFKRDFSYLTTINEGDLVPIYVDEVLPGDTIKIKQRSLV 59
Query 61 RMPSLLTPIMDNLYLDTYYFFVPNRIVWQHWKELMGENNESAWIPTTEYEVPQITAPSGG 120
RM + L P+MDN YLD +YFFVP R+VW HW+ LMGEN +S W P +Y P +APSGG
Sbjct 60 RMSTPLYPVMDNCYLDIWYFFVPCRLVWDHWQNLMGENTKSYWAPDVQYTTPLTSAPSGG 119
Query 121 WSIGTIADYMGVPTGVSGLSVNALPFRAYALICNEWFRDENLCDPLNIPLTDATVAGVNT 180
W +GTIADYMG+PTGVSG+ VN++P RAYA I NEWFRDENL P+ DAT G NT
Sbjct 120 WQVGTIADYMGIPTGVSGIKVNSMPMRAYARIWNEWFRDENLQQPVTQHSDDATTTGSNT 179
Query 181 GTFVTDVAKGGLPYKAAKYRDYFTSCLPAPQKSEDVTIPVSSGANYPVLSLSDIVPTPGT 240
GT +TD GGLP K AK++DYFTSCLPAPQK E + + + L + P
Sbjct 180 GTELTDAESGGLPLKVAKFKDYFTSCLPAPQKGEAIGFDFNQTPKVKGIGL--VFPLETN 237
Query 241 VPVKWNDANNVVSDAQWLLGGKNYNGTITSNDISLTKTNT------------GPTYSA-- 286
D DAQ L G+NYN + + + T+T GP SA
Sbjct 238 TGHTATDILWRQPDAQ--LVGENYNTSYNNFNSITTQTTVNGKKAFFFNNGKGPMLSARF 295
Query 287 -------VTPINLWAVNDGSVSSATINQLRLAFQVQKLYERDARGGTRYIEVLKSHFGVT 339
V + L AV + S + +IN LR A +Q + E DARGGTRY+E+LK+ FGV+
Sbjct 296 EDDYNGGVEQVELTAVAENSTNFLSINDLRQAIALQHILEADARGGTRYVEILKNEFGVS 355
Query 340 SPDARLQRPEYLGGNRIPIVISEINQTSGTSANSTPQGNPSGQSRTTDVHSDFKKSFVEH 399
SPDARLQR EY+GG RIPI +S++ Q+S + S PQGN + S TT ++ S VEH
Sbjct 356 SPDARLQRSEYIGGERIPINVSQVIQSSASDTTS-PQGNAAAYSLTTSANTIRAYSAVEH 414
Query 400 GFIIGVMVARYDHTYQQGLERFWSRKGRLDYYWPVFANIGEQAVLNKEIYAQGNGTDDEV 459
G+I+G+ R DH+YQQGL R W+R R YY P+ AN+GEQAVLN+EIYAQG D EV
Sbjct 415 GYILGLAAIRVDHSYQQGLSRMWTRSDRFSYYHPMLANLGEQAVLNQEIYAQGTTADTEV 474
Query 460 FGYQEAWADYRYKPNRVTGEMRSQAPQSLDVWHLGDDYSKLPSLSDSWVQEDSAVVNRVI 519
FGYQEAWADYRY+ N +TGEMRS QSLD WH GD Y+ LP LS+ W++E ++R +
Sbjct 475 FGYQEAWADYRYRTNMITGEMRSTYAQSLDAWHYGDKYTDLPRLSNDWIKEGQENIDRTL 534
Query 520 AVSEENSNQLWADIFIKNKCTRAMPMYSIPGL 551
AV ENS+Q +++ R MP+YS+PGL
Sbjct 535 AVQSENSHQFICNLYFDQTWVRPMPIYSVPGL 566
>gi|313766927|gb|ADR80653.1| putative major coat protein [Uncultured Microviridae]
Length=533
Score = 452 bits (1162), Expect = 2e-149, Method: Compositional matrix adjust.
Identities = 245/550 (45%), Positives = 340/550 (62%), Gaps = 44/550 (8%)
Query 5 TNSHFALNPTRIDMSRSTFDRNSSVKTSFNVGDIVPFYVDEVLPGDTFDIDTSKVIRMPS 64
T SH + D+ RSTF R +KT+FN GD++P YVDEVLPGDTF ++ + R+ +
Sbjct 12 TLSHEFSRVPQADIQRSTFSRVHGLKTTFNSGDLIPIYVDEVLPGDTFQMNATGFGRLAT 71
Query 65 LLTPIMDNLYLDTYYFFVPNRIVWQHWKELMGENNESAWIPTTEYEVPQITAPSGGWSIG 124
L P+MDN+Y++T++F+VPNRI+W +W++ G ++ +T++ VPQI S + G
Sbjct 72 PLYPVMDNMYVETFFFYVPNRIIWDNWEKFNGAQDDPN--DSTDFLVPQIQ--SATVAEG 127
Query 125 TIADYMGVPTGVSGLSVNALPFRAYALICNEWFRDENLCDPLNIPLTDATVAGVNTGTFV 184
++ DYMG+PT ++G+ N L RAY LI NEWFRDENL D L +P D
Sbjct 128 SLFDYMGLPTQIAGIDFNNLHGRAYNLIWNEWFRDENLQDSLGVPKDDGP---------- 177
Query 185 TDVAKGGLPYKAAKYRDYFTSCLPAPQKSEDVTIPVSSGANYPVLSLSDIVPTPGTVPVK 244
D G K K DYFTS LP PQK + V++P+ + A+
Sbjct 178 -DTYTGYTIQKRGKRHDYFTSALPWPQKGDAVSLPLGTSADI------------------ 218
Query 245 WNDANNVVSDAQWLLGGKNYNGTITSNDISLTKTNTGPTYSAVTPINLWAVNDGSVSSAT 304
+ A +D G + +TS+ + + + P + N + + ++AT
Sbjct 219 -HTAAAAGTDIGIYSVGSSDFRLLTSDPVEVALSGGTPPET-----NKMFADLSNATAAT 272
Query 305 INQLRLAFQVQKLYERDARGGTRYIEVLKSHFGVTSPDARLQRPEYLGGNRIPIVISEIN 364
INQLR AFQ+Q+LYE+DARGGTRY E+L+SHFGVTSPDARLQRPEYLGG + +++ +
Sbjct 273 INQLREAFQIQRLYEKDARGGTRYTEILQSHFGVTSPDARLQRPEYLGGQKTEVMMQTVP 332
Query 365 QTSGTSANSTPQGNPSGQSRTTDVHSDFKKSFVEHGFIIGVMVARYDHTYQQGLERFWSR 424
QTS T + S PQGN + T F KSFVEHG +IG+ D TYQQG+ R WSR
Sbjct 333 QTSSTDSTS-PQGNLAALGTATS-RGGFSKSFVEHGVLIGLACVFADLTYQQGMNRMWSR 390
Query 425 KGRLDYYWPVFANIGEQAVLNKEIYAQGNGTDDEVFGYQEAWADYRYKPNRVTGEMRSQA 484
+ R D+YWP A++GEQAVLN+EIY QG D + FGYQE +A+YRYKP+++TG+MRS A
Sbjct 391 RDRWDFYWPSLAHLGEQAVLNQEIYTQGTSADTQTFGYQERFAEYRYKPSQITGKMRSNA 450
Query 485 PQSLDVWHLGDDYSKLPSLSDSWVQEDSAVVNRVIAVSEENSNQLWADIFIKNKCTRAMP 544
+LD WHL D++ LP+L+ S+++E+ V+RVIAV E +W D + K TR MP
Sbjct 451 TGTLDAWHLAQDFTALPALNASFIEENPP-VDRVIAVPSE-PEFIW-DWYFDLKTTRPMP 507
Query 545 MYSIPGLIDH 554
+YS+PGLIDH
Sbjct 508 VYSVPGLIDH 517
>gi|530695351|gb|AGT39907.1| major capsid protein [Marine gokushovirus]
Length=539
Score = 450 bits (1157), Expect = 1e-148, Method: Compositional matrix adjust.
Identities = 245/569 (43%), Positives = 347/569 (61%), Gaps = 51/569 (9%)
Query 2 NRNTNSH-FALNPTRIDMSRSTFDRNSSVKTSFNVGDIVPFYVDEVLPGDTFDIDTSKVI 60
N++ ++H F++ P R ++ RS FD ++KT+F+ G +VP VDEVLPGD+ ++ +
Sbjct 5 NKSASAHQFSMIP-RAEIPRSKFDAQKTLKTAFDSGYLVPILVDEVLPGDSMNLRMTAFT 63
Query 61 RMPSLLTPIMDNLYLDTYYFFVPNRIVWQHWKELMGENNESAWIPTTEYEVPQITAPSGG 120
R+ + L P+MDN+YLDT++FFVPNR++W +W+ MGE + + +Y +P +T+P+GG
Sbjct 64 RLATPLFPVMDNMYLDTFFFFVPNRLLWSNWQRFMGERDPDP-DSSIDYTIPTMTSPNGG 122
Query 121 WSIGTIADYMGVPTGV-----SGLSVNALPFRAYALICNEWFRDENLCDPLNIPLTDATV 175
+++ ++ DYMG+PT S +S N+L RAY LI NEWFRDENL D + + D
Sbjct 123 YAVNSLQDYMGLPTAGQVDAGSSISHNSLFTRAYNLIWNEWFRDENLQDSVVVDKGD--- 179
Query 176 AGVNTGTFVTDVAKGGLPYKAAKYRDYFTSCLPAPQKSEDVTIPVSSGANYPVLSLSDIV 235
G +T T T + +G K DYFTS LP PQK + VT+P+ AN
Sbjct 180 -GPDTYTDYTLLRRG-------KRHDYFTSALPWPQKGDAVTLPLGGSAN---------- 221
Query 236 PTPGTVPVKWNDANNVVSDAQWLLGGKNYNGTITSNDISLTKTNTGPTYSAVTPINLWAV 295
V +ND D ++ N T + S++K G +N
Sbjct 222 -------VVYND----TGDPAYIREVSTGNVWTTPSRESVSKEANGNMSVPTGSVNAQYD 270
Query 296 NDGSV-------SSATINQLRLAFQVQKLYERDARGGTRYIEVLKSHFGVTSPDARLQRP 348
+GS+ ++ATIN +R +FQ+Q+L ERDARGGTRY E+++SHFGV SPDAR+QRP
Sbjct 271 PNGSLVADLSTATAATINAIRQSFQIQRLLERDARGGTRYTEIVRSHFGVISPDARMQRP 330
Query 349 EYLGGNRIPIVISEINQ--TSGTSANSTPQGNPSGQSRTTDVHSDFKKSFVEHGFIIGVM 406
EYLGG PI+++ + Q SG S TP G F SF EHG ++G+
Sbjct 331 EYLGGGSAPIIVNPVAQQSASGASGTDTPLGTLGAVGTGLASGHGFASSFTEHGVVVGLC 390
Query 407 VARYDHTYQQGLERFWSRKGRLDYYWPVFANIGEQAVLNKEIYAQGNGTDDEVFGYQEAW 466
R D TYQQGL R +SR R D+++PVF+++GEQ +LNKE+YA G TDD+VFGYQEAW
Sbjct 391 SVRADLTYQQGLHRMFSRSTRYDFFFPVFSHLGEQPILNKELYATGTSTDDDVFGYQEAW 450
Query 467 ADYRYKPNRVTGEMRSQAPQSLDVWHLGDDYSKLPSLSDSWVQEDSAVVNRVIAV-SEEN 525
A+YRYKP++VTG MRS A +LD WHL ++ LP+L+ +++ ED+ V+RV+AV SE N
Sbjct 451 AEYRYKPSQVTGLMRSTAAGTLDAWHLAQNFGSLPTLNSTFI-EDTPPVDRVVAVGSEAN 509
Query 526 SNQLWADIFIKNKCTRAMPMYSIPGLIDH 554
Q D F R MPMYS+PGL+DH
Sbjct 510 GQQFIFDAFFDINMARPMPMYSVPGLVDH 538
>gi|530695385|gb|AGT39938.1| major capsid protein [Marine gokushovirus]
Length=514
Score = 447 bits (1151), Expect = 5e-148, Method: Compositional matrix adjust.
Identities = 252/540 (47%), Positives = 337/540 (62%), Gaps = 47/540 (9%)
Query 15 RIDMSRSTFDRNSSVKTSFNVGDIVPFYVDEVLPGDTFDIDTSKVIRMPSLLTPIMDNLY 74
++D+ RS F+R+ +KT+F+ G +VP + DE LPGDTF +D + R+ + + P MDNLY
Sbjct 21 KVDIQRSVFNRDHGLKTTFDAGYLVPIFYDEALPGDTFTMDANGFGRLATPIAPFMDNLY 80
Query 75 LDTYYFFVPNRIVWQHWKELMGENNESAWIPTTEYEVPQITAPSGGWSIGTIADYMGVPT 134
++T++F VP R++W +W++ GE + +T+Y VPQ T G S T+ DY GVPT
Sbjct 81 IETFFFAVPYRLIWTNWEKFCGEQDNPG--DSTDYLVPQTT---GTISNSTLYDYFGVPT 135
Query 135 GVSGLSVNALPFRAYALICNEWFRDENLCDPLNIPLTDATVAGVNTGTFVTDVAKGGLPY 194
V+ L+ N L RAY L+ NEWFRD+NL + + + D G +T + T + +G
Sbjct 136 DVN-LTFNNLCGRAYNLVYNEWFRDQNLQNSVTVDKGD----GPDTASNYTLLKRG---- 186
Query 195 KAAKYRDYFTSCLPAPQKSEDVTIPVSSGANYPVLSLSDIVPTPGTVPVKWNDANNVVSD 254
K DYFTS LP PQK E VT+P+ + A P++S D TP N + S+
Sbjct 187 ---KRHDYFTSALPWPQKGEAVTLPLGTTA--PIMS-GDFTTTP---------TNYIPSN 231
Query 255 AQWLLGGKNYNGTITSNDISLTKTNTGPTYSAVTPINLWAVNDGSVSSATINQLRLAFQV 314
G N + D S T G +WA + ++ATINQLR AFQ+
Sbjct 232 ------GNNIPPQDANGDYSFAGTGVGG-------YGIWA-DLSDATAATINQLREAFQI 277
Query 315 QKLYERDARGGTRYIEVLKSHFGVTSPDARLQRPEYLGGNRIPIVISEINQTSGTSANST 374
Q+LYE+DARGGTRY EV++SHFGVTSPDARLQRPEYLGG + I I+ I QTS T A +T
Sbjct 278 QRLYEKDARGGTRYTEVIQSHFGVTSPDARLQRPEYLGGGKDRININPIAQTSSTDA-TT 336
Query 375 PQGNPSGQSRTTDVHSDFKKSFVEHGFIIGVMVARYDHTYQQGLERFWSRKGRLDYYWPV 434
PQGN SG T F KSF EH ++G+ D TYQQGL R +SR+ R D+YWP
Sbjct 337 PQGNLSGYGTTGFTGHRFNKSFTEHSVVLGLACVFADLTYQQGLPRHFSRQTRWDFYWPA 396
Query 435 FANIGEQAVLNKEIYAQGNGTDDEVFGYQEAWADYRYKPNRVTGEMRSQAPQSLDVWHLG 494
A++GEQAVLNKEIYAQG D+ VFGYQE +A+YRYKP+ +TG+MRS QSLD+WHL
Sbjct 397 LAHLGEQAVLNKEIYAQGTTDDNNVFGYQERYAEYRYKPSSITGQMRSNFAQSLDIWHLA 456
Query 495 DDYSKLPSLSDSWVQEDSAVVNRVIAVSEENSNQLWADIFIKNKCTRAMPMYSIPGLIDH 554
D+ LP L+ S+++E+ V+RV AV +N L D++ K KC R MP Y +PGLIDH
Sbjct 457 QDFGSLPVLNSSFIEENPP-VDRVTAV--QNYPNLILDMYFKLKCARPMPTYGVPGLIDH 513
Lambda K H a alpha
0.316 0.133 0.410 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 4045963415220