bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: All non-redundant GenBank CDS translations+PDB+SwissProt+PIR+PRF
excluding environmental samples from WGS projects
49,011,213 sequences; 17,563,301,199 total letters
Query= Contig-30_CDS_annotation_glimmer3.pl_2_3
Length=179
Score E
Sequences producing significant alignments: (Bits) Value
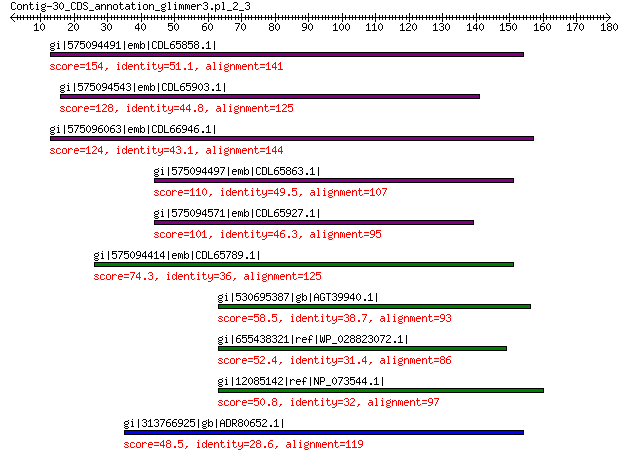
gi|575094491|emb|CDL65858.1| unnamed protein product 154 1e-43
gi|575094543|emb|CDL65903.1| unnamed protein product 128 6e-34
gi|575096063|emb|CDL66946.1| unnamed protein product 124 3e-32
gi|575094497|emb|CDL65863.1| unnamed protein product 110 8e-27
gi|575094571|emb|CDL65927.1| unnamed protein product 101 6e-24
gi|575094414|emb|CDL65789.1| unnamed protein product 74.3 2e-13
gi|530695387|gb|AGT39940.1| portal protein 58.5 5e-08
gi|655438321|ref|WP_028823072.1| hypothetical protein 52.4 7e-06
gi|12085142|ref|NP_073544.1| minor capsid protein 50.8 3e-05
gi|313766925|gb|ADR80652.1| putative minor capsid protein 48.5 2e-04
>gi|575094491|emb|CDL65858.1| unnamed protein product [uncultured bacterium]
Length=176
Score = 154 bits (389), Expect = 1e-43, Method: Compositional matrix adjust.
Identities = 72/141 (51%), Positives = 92/141 (65%), Gaps = 0/141 (0%)
Query 13 MPKFANQFEPHARIHTNPGTPEKVLYGPVFDENGTLDLEPKGKENLYDYIQSHKDSCDIK 72
M KF QF HAR+ PG P KV+Y P +DENG LDL+P G+ENLYD+IQSH S DI
Sbjct 1 MVKFKTQFHSHARVFQRPGDPIKVVYSPRYDENGVLDLQPTGEENLYDFIQSHAQSTDIH 60
Query 73 LIVDRCARGDLSALSKAQGMYGDFTTLPRTYAEALQALADAEHFFMRLPVETRAKFDHDP 132
+I+DR A G+ LS+ QG Y D + +P+TYAE L A+ E F RLPVE + KF +
Sbjct 61 VILDRFASGETDVLSQIQGFYADASDMPKTYAEVLNAVIAGEQTFDRLPVEIKQKFGNSF 120
Query 133 HKFIVSMDKPGFLEKLGVPVP 153
++ SMD P F E++G P P
Sbjct 121 STWLSSMDNPDFAERMGFPKP 141
>gi|575094543|emb|CDL65903.1| unnamed protein product [uncultured bacterium]
Length=145
Score = 128 bits (321), Expect = 6e-34, Method: Compositional matrix adjust.
Identities = 56/125 (45%), Positives = 86/125 (69%), Gaps = 0/125 (0%)
Query 16 FANQFEPHARIHTNPGTPEKVLYGPVFDENGTLDLEPKGKENLYDYIQSHKDSCDIKLIV 75
F +QF+ H R++ + G+P ++Y +D++G DL P G++N+YD IQSH+DS DI +++
Sbjct 2 FRSQFDDHDRVYCSSGSPIHIVYQGRYDDDGVFDLFPSGQDNIYDQIQSHRDSVDIHVLL 61
Query 76 DRCARGDLSALSKAQGMYGDFTTLPRTYAEALQALADAEHFFMRLPVETRAKFDHDPHKF 135
DR RGD+ LS QG+YGDFT +P +Y+E L A+ E FM LPV+ RAK+ H ++
Sbjct 62 DRYQRGDVDVLSARQGVYGDFTGMPASYSEILNAVLAGERAFMDLPVDERAKYGHSFAQW 121
Query 136 IVSMD 140
+ S+D
Sbjct 122 LSSLD 126
>gi|575096063|emb|CDL66946.1| unnamed protein product [uncultured bacterium]
Length=163
Score = 124 bits (311), Expect = 3e-32, Method: Compositional matrix adjust.
Identities = 62/145 (43%), Positives = 89/145 (61%), Gaps = 1/145 (1%)
Query 13 MPKFANQFEPHARIHTNPGTPEKVLYGPVFDENGTLDLEPKGKENLYDYIQSHKDSCDIK 72
M +F Q++PH RI T G+ E + YG +D+ G + LE G+ NLYD IQSH +S D+
Sbjct 1 MMEFKTQYDPHDRIFTEAGSREHITYGGHYDDEGRVVLEETGRINLYDEIQSHAESVDLH 60
Query 73 LIVDRCARGDLSALSKAQGMYGDFTTLPRTYAEALQALADAEHFFMRLPVETRAKFDHDP 132
+++ R + GD+ LS+ QG +GD P+TYAEAL + + E FM LP++ RAKF H
Sbjct 61 VLMQRYSCGDVDCLSQRQGFFGDVLDFPQTYAEALNHMQEMERQFMSLPLDVRAKFGHSF 120
Query 133 HKFIVSMDKPGFLEKLGV-PVPNSG 156
+F+ + FLE+LG P P S
Sbjct 121 SEFLAASGDDDFLERLGFKPEPESA 145
>gi|575094497|emb|CDL65863.1| unnamed protein product [uncultured bacterium]
Length=170
Score = 110 bits (274), Expect = 8e-27, Method: Compositional matrix adjust.
Identities = 53/107 (50%), Positives = 72/107 (67%), Gaps = 0/107 (0%)
Query 44 ENGTLDLEPKGKENLYDYIQSHKDSCDIKLIVDRCARGDLSALSKAQGMYGDFTTLPRTY 103
E+G +LE G+ +LY IQS+KDSCDI I++R ARGD SALSK QG+YGDFT +P
Sbjct 32 EDGKRELEKVGRIDLYAQIQSYKDSCDINYILERFARGDESALSKIQGVYGDFTAMPTNL 91
Query 104 AEALQALADAEHFFMRLPVETRAKFDHDPHKFIVSMDKPGFLEKLGV 150
AE Q + DAE F LPV+ RA+F+H P +F ++ F + +G+
Sbjct 92 AELQQRVVDAEALFYNLPVDIRAEFNHSPSEFYSAIGTDKFNKAVGI 138
>gi|575094571|emb|CDL65927.1| unnamed protein product [uncultured bacterium]
Length=133
Score = 101 bits (252), Expect = 6e-24, Method: Compositional matrix adjust.
Identities = 44/95 (46%), Positives = 64/95 (67%), Gaps = 0/95 (0%)
Query 44 ENGTLDLEPKGKENLYDYIQSHKDSCDIKLIVDRCARGDLSALSKAQGMYGDFTTLPRTY 103
++G +DL GK+NLYD IQSHKDS D+ L++ R G++ LS+ QG Y D + +P+TY
Sbjct 6 DDGAIDLVENGKKNLYDEIQSHKDSVDLNLLLQRFNNGEVDVLSRMQGTYADLSNMPKTY 65
Query 104 AEALQALADAEHFFMRLPVETRAKFDHDPHKFIVS 138
A+ L + E F+ LPV+ RAKFDH K++V+
Sbjct 66 ADMLNLIKKGEADFLSLPVDVRAKFDHSFEKWLVT 100
>gi|575094414|emb|CDL65789.1| unnamed protein product [uncultured bacterium]
Length=167
Score = 74.3 bits (181), Expect = 2e-13, Method: Compositional matrix adjust.
Identities = 45/125 (36%), Positives = 65/125 (52%), Gaps = 1/125 (1%)
Query 26 IHTNPGTPEKVLYGPVFDENGTLDLEPKGKENLYDYIQSHKDSCDIKLIVDRCARGDLSA 85
I PG + Y ENG LE G+ N Y IQS+KD CD+ I+ R GD S
Sbjct 16 IPNEPGNELEPHYIEKIGENGRTYLEKDGETNTYAEIQSYKDECDVHSILCRYFAGDTSV 75
Query 86 LSKAQGMYGDFTTLPRTYAEALQALADAEHFFMRLPVETRAKFDHDPHKFIVSMDKPGFL 145
LS+ QG+Y D T LP TY E +A+ F +LP+E + KFD+ + + + +
Sbjct 76 LSR-QGVYIDATQLPTTYHEMYNLMAEQRDKFDQLPLEIKRKFDNSFNVWASTAGSEEWY 134
Query 146 EKLGV 150
+ +G+
Sbjct 135 KLMGI 139
>gi|530695387|gb|AGT39940.1| portal protein [Marine gokushovirus]
Length=164
Score = 58.5 bits (140), Expect = 5e-08, Method: Compositional matrix adjust.
Identities = 36/103 (35%), Positives = 53/103 (51%), Gaps = 11/103 (11%)
Query 63 QSHKDSCDIKLIVDRCAR-GDLSALSKAQGMYGDFTTLPRTYAEALQALADAEHFFMRLP 121
Q KD C++ I+ + R G + + + Q YGDF+ + Y EAL + DA+ FM +P
Sbjct 36 QHFKDECEVINIIKKHDRNGIIEHVQRGQARYGDFSQVA-DYREALDLVRDAQQEFMSVP 94
Query 122 VETRAKFDHDPHKFIVSMDKP---------GFLEKLGVPVPNS 155
+ R KFD+DP KF + P GF+E V P+S
Sbjct 95 SDIRKKFDNDPGKFYEFVSNPDNKEELKQMGFIETPEVGKPSS 137
>gi|655438321|ref|WP_028823072.1| hypothetical protein [Proteobacteria bacterium JGI 0000113-P07]
Length=149
Score = 52.4 bits (124), Expect = 7e-06, Method: Compositional matrix adjust.
Identities = 27/87 (31%), Positives = 49/87 (56%), Gaps = 2/87 (2%)
Query 63 QSHKDSCDIKLIVDRCAR-GDLSALSKAQGMYGDFTTLPRTYAEALQALADAEHFFMRLP 121
Q ++ CDI I+ R R G + + + Q YGDF+ + Y EAL + +A FM++P
Sbjct 31 QHFQEECDIINIIKRHDRNGIIEHVHRGQARYGDFSEV-HDYREALDLVQEANEEFMKIP 89
Query 122 VETRAKFDHDPHKFIVSMDKPGFLEKL 148
+ R +F+++P +F+ + P E++
Sbjct 90 SDIRKQFNNNPGEFLNFVSNPDNEEEI 116
>gi|12085142|ref|NP_073544.1| minor capsid protein [Bdellovibrio phage phiMH2K]
gi|75089167|sp|Q9G053.1|B_BPPHM RecName: Full=Internal scaffolding protein VP3 [Bdellovibrio
phage phiMH2K]
gi|12017990|gb|AAG45346.1|AF306496_7 Vp3 [Bdellovibrio phage phiMH2K]
Length=151
Score = 50.8 bits (120), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 31/109 (28%), Positives = 51/109 (47%), Gaps = 13/109 (12%)
Query 63 QSHKDSCDIKLIVDRCAR-GDLSALSKAQGMYGDFTTLPRTYAEALQALADAEHFFMRLP 121
QS CDI IV + + G ++ L++ QG YGD +++P + A+ + A+ F LP
Sbjct 26 QSFGKECDINFIVKKFIKTGQITHLARRQGFYGDQSSIPD-FQTAMDTVTKAQQAFDELP 84
Query 122 VETRAKFDHDPHKFIVSMDKP---------GFLEKLGVP--VPNSGAEQ 159
R +F + PH+ + + P G +E + P P S Q
Sbjct 85 AHMRKRFANSPHELMQFLQDPKNRDEAISLGLMEMVETPQQAPQSTTNQ 133
>gi|313766925|gb|ADR80652.1| putative minor capsid protein [Uncultured Microviridae]
Length=153
Score = 48.5 bits (114), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 34/120 (28%), Positives = 57/120 (48%), Gaps = 17/120 (14%)
Query 35 KVLYGPVFDENGTLDLEPKGKENLYDYIQSHKDSCDIKLIVDRCARGDLSA-LSKAQGMY 93
K LY PV+++ T QS KDSCDI I+ R + ++A L++ + Y
Sbjct 9 KELYEPVYEDGRTK--------------QSFKDSCDINKILARFSGEQVAAHLTQYKNEY 54
Query 94 GDFTTLPRTYAEALQALADAEHFFMRLPVETRAKFDHDPHKFIVSMDKPGFLEKLGVPVP 153
GDF+ + L LA F +LP+ + +F++ P +F ++ E+L +P
Sbjct 55 GDFSEFDFQKNQJL--LARGNQIFDKLPISVKREFNNQPSEFFTFVNDSNNSERLAELLP 112
Lambda K H a alpha
0.318 0.136 0.406 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 442789906005