bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: All non-redundant GenBank CDS translations+PDB+SwissProt+PIR+PRF
excluding environmental samples from WGS projects
49,011,213 sequences; 17,563,301,199 total letters
Query= Contig-30_CDS_annotation_glimmer3.pl_2_5
Length=77
Score E
Sequences producing significant alignments: (Bits) Value
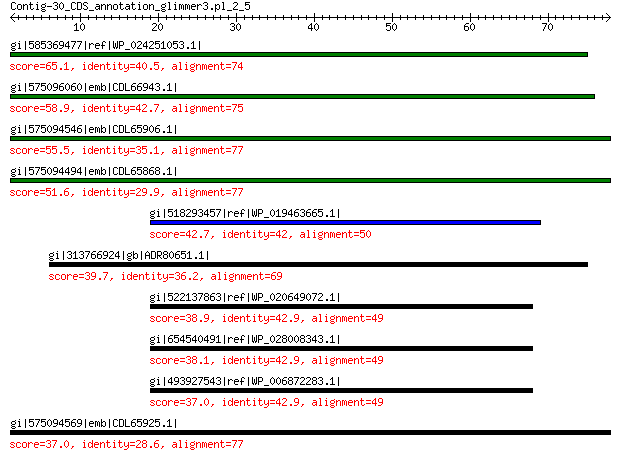
gi|585369477|ref|WP_024251053.1| hypothetical protein 65.1 5e-11
gi|575096060|emb|CDL66943.1| unnamed protein product 58.9 2e-08
gi|575094546|emb|CDL65906.1| unnamed protein product 55.5 4e-07
gi|575094494|emb|CDL65868.1| unnamed protein product 51.6 7e-06
gi|518293457|ref|WP_019463665.1| hypothetical protein 42.7 0.009
gi|313766924|gb|ADR80651.1| putative replication initiation protein 39.7 0.073
gi|522137863|ref|WP_020649072.1| hypothetical protein 38.9 0.18
gi|654540491|ref|WP_028008343.1| RND transporter MFP subunit 38.1 0.30
gi|493927543|ref|WP_006872283.1| hypothetical protein 37.0 0.57
gi|575094569|emb|CDL65925.1| unnamed protein product 37.0 0.78
>gi|585369477|ref|WP_024251053.1| hypothetical protein [Escherichia coli]
Length=243
Score = 65.1 bits (157), Expect = 5e-11, Method: Composition-based stats.
Identities = 30/74 (41%), Positives = 48/74 (65%), Gaps = 0/74 (0%)
Query 1 VSTRQGGVKFRPPRYFNRLLETDNPELSEKLREARKNLAKEISRCKLSQTDKRYMNMLDT 60
+ST GG K RPP+YF++L + + PEL +++ RK+ A+E + KL+Q+ Y +L+T
Sbjct 168 ISTPDGGRKIRPPKYFDKLFDLEQPELMAEIKAKRKHFAEEGKKAKLAQSTMTYEEILET 227
Query 61 EEEVKQEAVKTLRR 74
+E V +K LRR
Sbjct 228 QERVLHNRIKNLRR 241
>gi|575096060|emb|CDL66943.1| unnamed protein product [uncultured bacterium]
Length=339
Score = 58.9 bits (141), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 32/75 (43%), Positives = 44/75 (59%), Gaps = 0/75 (0%)
Query 1 VSTRQGGVKFRPPRYFNRLLETDNPELSEKLREARKNLAKEISRCKLSQTDKRYMNMLDT 60
+ST +GG KFRPPRYF +L E D PE + K E RK KL++T+ ++ML
Sbjct 263 ISTLKGGRKFRPPRYFEKLFELDFPEEAAKRSEVRKAAGSNAMAAKLAKTNLDPLSMLAV 322
Query 61 EEEVKQEAVKTLRRS 75
EE + +K LRR+
Sbjct 323 EERNFTDRIKPLRRN 337
>gi|575094546|emb|CDL65906.1| unnamed protein product [uncultured bacterium]
Length=351
Score = 55.5 bits (132), Expect = 4e-07, Method: Composition-based stats.
Identities = 27/77 (35%), Positives = 41/77 (53%), Gaps = 0/77 (0%)
Query 1 VSTRQGGVKFRPPRYFNRLLETDNPELSEKLREARKNLAKEISRCKLSQTDKRYMNMLDT 60
+ST +GG+ PP+YF+ E D P+ + R LA+E KL Q+ Y ML
Sbjct 275 ISTERGGLSMLPPKYFDHFYELDAPDDYVNYKSVRAALARESLLLKLDQSTLSYGQMLAV 334
Query 61 EEEVKQEAVKTLRRSKL 77
E +K +K+L RS++
Sbjct 335 AESIKNNKIKSLDRSEV 351
>gi|575094494|emb|CDL65868.1| unnamed protein product [uncultured bacterium]
Length=348
Score = 51.6 bits (122), Expect = 7e-06, Method: Composition-based stats.
Identities = 23/77 (30%), Positives = 46/77 (60%), Gaps = 0/77 (0%)
Query 1 VSTRQGGVKFRPPRYFNRLLETDNPELSEKLREARKNLAKEISRCKLSQTDKRYMNMLDT 60
+ T++ ++ +PP+YF +LLE +N ++ ++ R+ +LA++ S + T Y+ ML
Sbjct 272 LGTKEKSIQMKPPKYFEKLLEKENEDVFKERRDLHASLAEDFSCLRNLSTSHDYLGMLQM 331
Query 61 EEEVKQEAVKTLRRSKL 77
EE+ +KTL+R +
Sbjct 332 EEDNLNARIKTLKRKEF 348
>gi|518293457|ref|WP_019463665.1| hypothetical protein [Dyella japonica]
gi|664785830|gb|AIF48597.1| RND transporter MFP subunit [Dyella japonica A8]
Length=389
Score = 42.7 bits (99), Expect = 0.009, Method: Composition-based stats.
Identities = 21/51 (41%), Positives = 32/51 (63%), Gaps = 1/51 (2%)
Query 19 LLETDNPELSEKLREARKNLAKEISRCKLS-QTDKRYMNMLDTEEEVKQEA 68
L E D PEL ++ +A+ NLA+ + +L+ TDKR+ N+L + KQEA
Sbjct 110 LAEIDTPELDQQFEQAKANLARAQANARLAVLTDKRWKNLLTSNSVSKQEA 160
>gi|313766924|gb|ADR80651.1| putative replication initiation protein [Uncultured Microviridae]
Length=285
Score = 39.7 bits (91), Expect = 0.073, Method: Composition-based stats.
Identities = 25/69 (36%), Positives = 40/69 (58%), Gaps = 7/69 (10%)
Query 6 GGVKFRPPRYFNRLLETDNPELSEKLREARKNLAKEISRCKLSQTDKRYMNMLDTEEEVK 65
GGVK PR++++L+E ++PE E ++E RK A + + D +LD + VK
Sbjct 222 GGVKNGIPRFYDKLMEKEDPEQLEIVKEKRKEFALD------NMHDNTGPRLLD-KATVK 274
Query 66 QEAVKTLRR 74
A+KTL+R
Sbjct 275 LAAIKTLKR 283
>gi|522137863|ref|WP_020649072.1| hypothetical protein [Solimonas variicoloris]
Length=395
Score = 38.9 bits (89), Expect = 0.18, Method: Composition-based stats.
Identities = 21/50 (42%), Positives = 33/50 (66%), Gaps = 1/50 (2%)
Query 19 LLETDNPELSEKLREARKNLAKEISRCKLSQ-TDKRYMNMLDTEEEVKQE 67
L E + PEL ++L +AR +LAK + +L++ T KR+ +ML T+ KQE
Sbjct 108 LAEIETPELDQQLAQARADLAKAEADAELARSTAKRWQSMLGTDAVSKQE 157
>gi|654540491|ref|WP_028008343.1| RND transporter MFP subunit [Solimonas flava]
Length=394
Score = 38.1 bits (87), Expect = 0.30, Method: Composition-based stats.
Identities = 21/50 (42%), Positives = 33/50 (66%), Gaps = 1/50 (2%)
Query 19 LLETDNPELSEKLREARKNLAKEISRCKLSQ-TDKRYMNMLDTEEEVKQE 67
L E + PEL ++L +AR +LAK + +L++ T KR+ +ML T+ KQE
Sbjct 108 LAEIETPELDQQLAQARADLAKAEADAELARSTAKRWQSMLGTDAVSKQE 157
>gi|493927543|ref|WP_006872283.1| hypothetical protein [Legionella drancourtii]
gi|363536000|gb|EHL29446.1| hypothetical protein LDG_8407 [Legionella drancourtii LLAP12]
Length=287
Score = 37.0 bits (84), Expect = 0.57, Method: Composition-based stats.
Identities = 21/50 (42%), Positives = 32/50 (64%), Gaps = 1/50 (2%)
Query 19 LLETDNPELSEKLREARKNLAKEISRCKLSQ-TDKRYMNMLDTEEEVKQE 67
L E + PEL+ + R+A +L I+ KL+Q T KR++N+L T+ KQE
Sbjct 13 LAEIETPELNAQKRQAAADLNTAIANNKLAQSTAKRWINLLKTDSVSKQE 62
>gi|575094569|emb|CDL65925.1| unnamed protein product [uncultured bacterium]
Length=354
Score = 37.0 bits (84), Expect = 0.78, Method: Composition-based stats.
Identities = 22/77 (29%), Positives = 38/77 (49%), Gaps = 0/77 (0%)
Query 1 VSTRQGGVKFRPPRYFNRLLETDNPELSEKLREARKNLAKEISRCKLSQTDKRYMNMLDT 60
+ST +GG+ + P YF RL+E + EL ++++ + K A + TD Y+ L
Sbjct 277 LSTLKGGMSMQIPPYFIRLIEDIDSELFKEIKRSNKQAALNHQEALMKNTDVDYITYLSF 336
Query 61 EEEVKQEAVKTLRRSKL 77
E + K RR ++
Sbjct 337 LEGILVREEKFYRRDRI 353
Lambda K H a alpha
0.313 0.129 0.346 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 441112126275