bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: All non-redundant GenBank CDS translations+PDB+SwissProt+PIR+PRF
excluding environmental samples from WGS projects
49,011,213 sequences; 17,563,301,199 total letters
Query= Contig-31_CDS_annotation_glimmer3.pl_2_1
Length=534
Score E
Sequences producing significant alignments: (Bits) Value
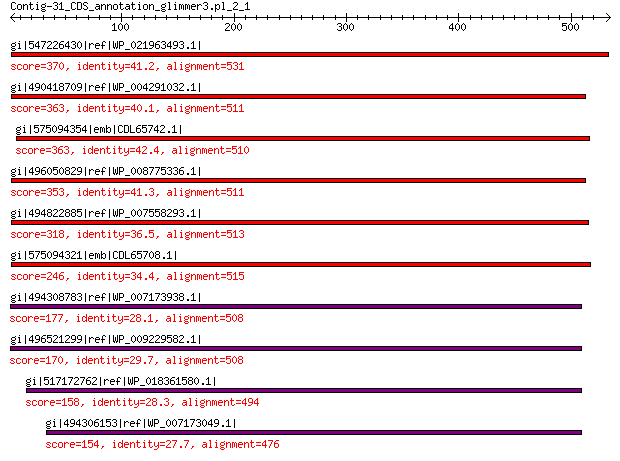
gi|547226430|ref|WP_021963493.1| putative uncharacterized protein 370 1e-117
gi|490418709|ref|WP_004291032.1| hypothetical protein 363 1e-114
gi|575094354|emb|CDL65742.1| unnamed protein product 363 4e-114
gi|496050829|ref|WP_008775336.1| hypothetical protein 353 6e-111
gi|494822885|ref|WP_007558293.1| hypothetical protein 318 6e-97
gi|575094321|emb|CDL65708.1| unnamed protein product 246 2e-69
gi|494308783|ref|WP_007173938.1| hypothetical protein 177 5e-45
gi|496521299|ref|WP_009229582.1| capsid protein 170 9e-43
gi|517172762|ref|WP_018361580.1| hypothetical protein 158 1e-38
gi|494306153|ref|WP_007173049.1| hypothetical protein 154 3e-37
>gi|547226430|ref|WP_021963493.1| putative uncharacterized protein [Prevotella sp. CAG:1185]
gi|524103382|emb|CCY83994.1| putative uncharacterized protein [Prevotella sp. CAG:1185]
Length=573
Score = 370 bits (951), Expect = 1e-117, Method: Compositional matrix adjust.
Identities = 219/551 (40%), Positives = 317/551 (58%), Gaps = 55/551 (10%)
Query 2 SLFNLSSVKNHPRRSGFDLSSKVAFTAKVGELLPVKWTLTMPGDKFSLKEQHFTRTQPVN 61
S+ +L+++KN +R+GFDLS K AFTAKVGELLP+ PGDKF+++ Q FTRTQPVN
Sbjct 3 SVMSLTALKNSVKRNGFDLSFKNAFTAKVGELLPIMCKEVYPGDKFNIRGQAFTRTQPVN 62
Query 62 TSAYTRVREYYDWFWCPLHLLWRNAPEVISQIQQNVQHASSFDGSVLLGSNMPCLSADQI 121
++AY+R+REYYD+++ P LLW AP + + + HA+ SV L P + I
Sbjct 63 SAAYSRLREYYDFYFVPYRLLWNMAPTFFTNM-PDPHHAADLVSSVNLSQRHPWFTFFDI 121
Query 122 SQSLDQLKS--------KQNYFGFDRADLAYKLLQYLRYGNVRTGVGSNGARNYGTSIDV 173
+ L L S ++N+FGF R +L+ KLL YL YG G + S D+
Sbjct 122 MEYLGNLNSLSGAYEKYQKNFFGFSRVELSVKLLNYLNYG---FGKDYESVKVPSDSDDI 178
Query 174 KDGTYNQNRAYNHALSIFPILAYKKFCQDYFRLTQWQDSAPYLWNIDYYDGKGAVTILPT 233
LS FP+LAY+K C+DYFR QWQ +APY +N+DY GK + +P
Sbjct 179 -------------VLSPFPLLAYQKICEDYFRDDQWQSAAPYRYNLDYLYGKSSGFHIPM 225
Query 234 SLTSSAAYFEGDTFFDLEYCNWNKDMFFGILPDAQFGDTSVVDISYGVTGAPVVTKQNLQ 293
S ++ A F+ T FDL YCN+ KD F G+LP AQ+GD SV +G +L
Sbjct 226 SSFTNDA-FKNPTMFDLNYCNFQKDYFTGMLPRAQYGDVSVASPIFG----------DLD 274
Query 294 SPTNSSVSIGTDDANSKTLIASG---------TNLTLDVLALRRGEALQRFREISLCTPL 344
+SS++ + I SG T L VLALR+ E LQ++REI+ +
Sbjct 275 IGDSSSLTFASAPQQGANTIQSGVLVVNNNSNTTAGLSVLALRQAECLQKWREIAQSGKM 334
Query 345 NYRSQIKAHFGVDVGAAMSGMSTYIGGEASSLDISEVVNTNITETNEALIAGKGVGTGQS 404
+Y++Q++ HF V A +SG Y+GG S+LDISEVVNTN+T N+A I GKG GT
Sbjct 335 DYQTQMQKHFNVSPSATLSGHCKYLGGWTSNLDISEVVNTNLTGDNQADIQGKGTGTLNG 394
Query 405 SE-SFYAKDWGILMCIYHSVPLLDYVLSAPDPQLFTSENTSFPVPELDSIGLEPISVAYY 463
++ F + + GI+MCIYH +PLLD+ ++ Q F + T + +PE DS+G++ +
Sbjct 395 NKVDFESSEHGIIMCIYHCLPLLDWSINRIARQNFKTTFTDYAIPEFDSVGMQQLY---- 450
Query 464 SNNPTELP-SASGLLSDP-TVTVGYLPRYFAWKTSLDYVLGAFTTTEKEWVAPITQTLWT 521
P+E+ L SDP ++ +GY+PRY KTS+D + G+F T WV+P+T + +
Sbjct 451 ---PSEMIFGLEDLPSDPSSINMGYVPRYADLKTSIDEIHGSFIDTLVSWVSPLTDSYIS 507
Query 522 KYVEAFERFGY 532
Y +A + G+
Sbjct 508 AYRQACKDAGF 518
>gi|490418709|ref|WP_004291032.1| hypothetical protein [Bacteroides eggerthii]
gi|217986636|gb|EEC52970.1| putative capsid protein (F protein) [Bacteroides eggerthii DSM
20697]
Length=578
Score = 363 bits (932), Expect = 1e-114, Method: Compositional matrix adjust.
Identities = 205/525 (39%), Positives = 306/525 (58%), Gaps = 46/525 (9%)
Query 2 SLFNLSSVKNHPRRSGFDLSSKVAFTAKVGELLPVKWTLTMPGDKFSLKEQHFTRTQPVN 61
++ +L S++N P R+GFDLS K FTAK GELLPV +PGD F + + FTRTQPVN
Sbjct 3 NIMSLKSIRNKPSRNGFDLSFKKNFTAKAGELLPVMVKEVLPGDTFKINLKAFTRTQPVN 62
Query 62 TSAYTRVREYYDWFWCPLHLLWRNAPEVISQIQQNVQHASSFDGS--VLLGSNMPCLSAD 119
T+A+ R+REYYD+F+ P LLW A V++Q+ N QHA S D + +L MP ++++
Sbjct 63 TAAFARIREYYDFFFVPYDLLWNKANTVLTQMYDNPQHAVSIDPTRNFVLSGEMPYMTSE 122
Query 120 QISQSLDQLKS-------KQNYFGFDRADLAYKLLQYLRYGNVRTGVGSNGARNYGTSID 172
I+ ++ L + K NYFG++R+ + KLL+YL YGN + + ++ T+
Sbjct 123 AIASYINALSTASALADYKSNYFGYNRSKSSVKLLEYLGYGNYESFL----TDDWNTAPL 178
Query 173 VKDGTYNQNRAYNHALSIFPILAYKKFCQDYFRLTQWQDSAPYLWNIDYYDGKGAVTILP 232
+ + +N IF +LAY+K D++R +QW+ +P +N+DY DG +
Sbjct 179 MANLNHN----------IFGLLAYQKIYSDFYRDSQWERVSPSTFNVDYLDGSS----MN 224
Query 233 TSLTSSAAYFEGDTFFDLEYCNWNKDMFFGILPDAQFGDTSVVDISYGVTGAPVVTKQNL 292
S +++ FFDL YCNW KD+F G+LP Q+G+T+V I+ VTG +T N
Sbjct 225 LDNAYSTEFYQNYNFFDLRYCNWQKDLFHGVLPHQQYGETAVASITPDVTGK--LTLSNF 282
Query 293 Q----SPTNSSVSIGTDDANSKTLIASGTNLTLDVLALRRGEALQRFREISLCTPLNYRS 348
SPT +S GT +K L A T L +L LR+ E LQ+++EI+ +Y+
Sbjct 283 STVGTSPTTAS---GT---ATKNLPAFDTVGDLSILVLRQAEFLQKWKEITQSGNKDYKD 336
Query 349 QIKAHFGVDVGAAMSGMSTYIGGEASSLDISEVVNTNITETNEALIAGKGVGTGQSSESF 408
Q++ H+GV VG S + TY+GG +SS+DI+EV+NTNIT + A IAGKGVG +F
Sbjct 337 QLEKHWGVSVGDGFSELCTYLGGVSSSIDINEVINTNITGSAAADIAGKGVGVANGEINF 396
Query 409 YAKD-WGILMCIYHSVPLLDYVLSAPDPQLFTSENTSFPVPELDSIGLEPISVAYYSNNP 467
+ +G++MCIYH +PLLDY DP +T + +PE D +G++ + + N
Sbjct 397 NSNGRYGLIMCIYHCLPLLDYTTDMLDPAFLKVNSTDYAIPEFDRVGMQSMPLVQLMNPL 456
Query 468 TELPSASGLLSDPTVTVGYLPRYFAWKTSLDYVLGAFTTTEKEWV 512
+ASGL+ +GY+PRY +KTS+D +G F T WV
Sbjct 457 RSFANASGLV------LGYVPRYIDYKTSVDQSVGGFKRTLNSWV 495
>gi|575094354|emb|CDL65742.1| unnamed protein product [uncultured bacterium]
Length=615
Score = 363 bits (931), Expect = 4e-114, Method: Compositional matrix adjust.
Identities = 216/564 (38%), Positives = 317/564 (56%), Gaps = 79/564 (14%)
Query 6 LSSVKNHPRRSGFDLSSKVAFTAKVGELLPVKWTLTMPGDKFSLKEQHFTRTQPVNTSAY 65
++ +KN P R+GFDLS K FTAK GELLPV + +PGD F++ + FTRTQP+NTSA+
Sbjct 3 MADIKNRPSRNGFDLSFKKNFTAKAGELLPVMTKVVLPGDSFNINLRSFTRTQPLNTSAF 62
Query 66 TRVREYYDWFWCPLHLLWRNAPEVISQIQQNVQHAS--SFDGSVLLGSNMPCLSADQISQ 123
R+REYYD+++ P +W I+Q+ NVQHAS + D + L MP +++QI+
Sbjct 63 ARMREYYDFYFVPFEQMWNKFDSCITQMNANVQHASGPTLDDNTPLSGRMPYFTSEQIAD 122
Query 124 SLDQ--LKSKQNYFGFDRADLAYKLLQYLRYGNVRTGVGSNGARNYGTSIDVKDGTYN-Q 180
L+ +++N FGF+R+ L KLLQYL YG+ S D + T++ +
Sbjct 123 YLNDQATAARKNPFGFNRSTLTCKLLQYLGYGDYN-------------SFDSETNTWSAK 169
Query 181 NRAYNHALSIFPILAYKKFCQDYFRLTQWQDSAPYLWNIDYYDGKGAVTILPTSLTSSAA 240
YN LS FP+LAY+K D++R TQW+ + P +N+DY G + + T L S
Sbjct 170 PLLYNLELSPFPLLAYQKIYSDFYRYTQWEKTNPSTFNLDYIKGTSDLQMDLTGLPS--- 226
Query 241 YFEGDTFFDLEYCNWNKDMFFGILPDAQFGDTSVVD-------ISYGVTGAPVVTKQ-NL 292
+ + FFD+ YCN+ KDMF G+LP AQ+G SVV IS G +G T +
Sbjct 227 --DDNNFFDIRYCNYQKDMFHGVLPVAQYGSASVVPINGQLNVISNGDSGPIFKTSTPDP 284
Query 293 QSPTNSSVSIGTD------------------------------DANSKTLIASGTNLTLD 322
+P S V++G + +A++++L+ NL ++
Sbjct 285 GTPGTSYVTVGGNIGVDNRSFGVSGSTLNVGKSADPSGYGFPSNASTRSLLWENPNLIIE 344
Query 323 --------VLALRRGEALQRFREISLCTPLNYRSQIKAHFGVDVGAAMSGMSTYIGGEAS 374
+LALR+ E LQ+++E+S+ +Y+SQI+ H+G+ V +S + Y+GG A+
Sbjct 345 NNQGFYVPILALRQAEFLQKWKEVSVSGEEDYKSQIEKHWGIKVSDFLSHQARYLGGCAT 404
Query 375 SLDISEVVNTNITETNEALIAGKGVGTGQSSESFYAK-DWGILMCIYHSVPLLDYVLSAP 433
SLDI+EV+N NIT N A IAGKG TG S F +K ++GI+MCIYH +P++DYV S
Sbjct 405 SLDINEVINNNITGDNAADIAGKGTFTGNGSIRFESKGEYGIIMCIYHVLPIVDYVGSGV 464
Query 434 DPQLFTSENTSFPVPELDSIGLE--PISVAYYSNNPTELPSASGLLSDPTVTVGYLPRYF 491
D + TSFP+PELD IG+E P+ A ++ PSA L GY PRY
Sbjct 465 DHSCTLVDATSFPIPELDQIGMESVPLVRAMNPVKESDTPSADTFL-------GYAPRYI 517
Query 492 AWKTSLDYVLGAFTTTEKEWVAPI 515
WKTS+D +G F + + W P+
Sbjct 518 DWKTSVDRSVGDFADSLRTWCLPV 541
>gi|496050829|ref|WP_008775336.1| hypothetical protein [Bacteroides sp. 2_2_4]
gi|229448893|gb|EEO54684.1| putative capsid protein (F protein) [Bacteroides sp. 2_2_4]
Length=580
Score = 353 bits (907), Expect = 6e-111, Method: Compositional matrix adjust.
Identities = 211/527 (40%), Positives = 310/527 (59%), Gaps = 41/527 (8%)
Query 2 SLFNLSSVKNHPRRSGFDLSSKVAFTAKVGELLPVKWTLTMPGDKFSLKEQHFTRTQPVN 61
++ +L S++N R+GFDLSSK FTAK GELLPVK +PGDK+S+ + FTRTQP+N
Sbjct 3 NIMSLKSLRNKTSRNGFDLSSKRNFTAKPGELLPVKCWEVLPGDKWSIDLKSFTRTQPLN 62
Query 62 TSAYTRVREYYDWFWCPLHLLWRNAPEVISQIQQNVQHASSFDGSV--LLGSNMPCLSAD 119
T+A+ R+REYYD+++ P +LLW A V++Q+ N QHA+S+ S L MP ++
Sbjct 63 TAAFARMREYYDFYFVPYNLLWNKANTVLTQMYDNPQHATSYIPSANQALAGVMPNVTCK 122
Query 120 QISQSLDQLKS--------KQNYFGFDRADLAYKLLQYLRYGNVRTGVGSNGARNYGTSI 171
I+ L+ + ++NYFG+ R+ KLL+YL YGN T Y TS
Sbjct 123 GIADYLNLVAPDVTTTNSYEKNYFGYSRSLGTAKLLEYLGYGNFYT---------YATS- 172
Query 172 DVKDGTYNQN-RAYNHALSIFPILAYKKFCQDYFRLTQWQDSAPYLWNIDYYDGKGAVTI 230
K+ T+ ++ + N L+I+ +LAY+K D+ R +QW+ +P +N+DY G +
Sbjct 173 --KNNTWTKSPLSSNLQLNIYGVLAYQKIYADHIRDSQWEKVSPSCFNVDYLSGTVDSAM 230
Query 231 LPTSLTSSAAYFEGDTFFDLEYCNWNKDMFFGILPDAQFGDTSVVDISYGVTGAPVVTKQ 290
S+ + + FDL YCNW KD+F G+LP Q+GDT+ V+++ + V++ Q
Sbjct 231 TIDSMITGQGFAPFYNMFDLRYCNWQKDLFHGVLPRQQYGDTAAVNVNL----SNVLSAQ 286
Query 291 NL-QSPTNSSVS---IGTDDANSKTLIASGTNLTLDVLALRRGEALQRFREISLCTPLNY 346
+ Q+P V + N +T+ SG T VLALR+ E LQ+++EI+ +Y
Sbjct 287 YMVQTPDGDPVGGSPFSSTGVNLQTVNGSG---TFTVLALRQAEFLQKWKEITQSGNKDY 343
Query 347 RSQIKAHFGVDVGAAMSGMSTYIGGEASSLDISEVVNTNITETNEALIAGKGVGTGQSSE 406
+ QI+ H+ V VG A S MS Y+GG +SLDI+EVVN NIT +N A IAGKGV G
Sbjct 344 KDQIEKHWNVSVGEAYSEMSLYLGGTTASLDINEVVNNNITGSNAADIAGKGVVVGNGRI 403
Query 407 SFYAKD-WGILMCIYHSVPLLDYVLSAPDPQLFTSENTSFPVPELDSIGLEPISVAYYSN 465
SF A + +G++MCIYHS+PLLDY +P +T F +PE D +G+E + + N
Sbjct 404 SFDAGERYGLIMCIYHSLPLLDYTTDLVNPAFTKINSTDFAIPEFDRVGMESVPLVSLMN 463
Query 466 NPTELPSASGLLSDPTVTVGYLPRYFAWKTSLDYVLGAFTTTEKEWV 512
L S+ + S +GY PRY ++KT +D +GAF TT K WV
Sbjct 464 ---PLQSSYNVGSS---ILGYAPRYISYKTDVDSSVGAFKTTLKSWV 504
>gi|494822885|ref|WP_007558293.1| hypothetical protein [Bacteroides plebeius]
gi|198272099|gb|EDY96368.1| putative capsid protein (F protein) [Bacteroides plebeius DSM
17135]
Length=613
Score = 318 bits (815), Expect = 6e-97, Method: Compositional matrix adjust.
Identities = 187/546 (34%), Positives = 286/546 (52%), Gaps = 50/546 (9%)
Query 2 SLFNLSSVKNHPRRSGFDLSSKVAFTAKVGELLPVKWTLTMPGDKFSLKEQHFTRTQPVN 61
++ ++ SV+N P R+G+DL+ K+ FTAK G L+PV WT +P D + + F RTQP+N
Sbjct 10 NIMSMKSVRNKPTRAGYDLTQKINFTAKAGSLIPVWWTPVLPFDDLNATVKSFVRTQPLN 69
Query 62 TSAYTRVREYYDWFWCPLHLLWRNAPEVISQIQQNVQHASS--FDGSVLLGSNMPCLSAD 119
T+A+ R+R Y+D+++ P +W P I+Q++ N+ HAS +V L +P +A+
Sbjct 70 TAAFARMRGYFDFYFVPFRQMWNKFPTAITQMRTNLLHASGPVLADNVPLSDELPYFTAE 129
Query 120 QISQSLDQLKSKQNYFGFDRADLAYKLLQYLRYGN----VRTGVGSNGARNYGTSIDVKD 175
Q++ + L +N FG+ RA L +L+YL YG+ + G GA
Sbjct 130 QVADYIVSLADSKNQFGYYRAWLVCIILEYLGYGDFYPYIVEAAGGEGA----------- 178
Query 176 GTYNQNRAYNH-ALSIFPILAYKKFCQDYFRLTQWQDSAPYLWNIDYYDGKGAVTILPTS 234
T+ N+ S FP+ AY+K D+ R TQW+ S P +NIDY G L +
Sbjct 179 -TWATRPMLNNLKFSPFPLFAYQKIYADFNRYTQWERSNPSTFNIDYISGSADSLQLDFT 237
Query 235 LTSSAAYFEGDTFFDLEYCNWNKDMFFGILPDAQFGDTSVVDIS------YGVTGAPVVT 288
+ F FD+ Y NW +D+ G +P AQ+G+ S V +S G T T
Sbjct 238 VEGFKDSF---NLFDMRYSNWQRDLLHGTIPQAQYGEASAVPVSGSMQVVEGPTPPAFTT 294
Query 289 KQNLQSPTNSSVSIGT-------------------DDANSKTLIASGTNLTLDVLALRRG 329
Q+ + N +V+I ++ NS ++ ++ + +LALRR
Sbjct 295 GQDGVAFLNGNVTIQGSSGYLQAQTSVGESRILRFNNTNSGLIVEGDSSFGVSILALRRA 354
Query 330 EALQRFREISLCTPLNYRSQIKAHFGVDVGAAMSGMSTYIGGEASSLDISEVVNTNITET 389
EA Q+++E++L + +Y SQI+AH+G V A S M ++G L I+EVVN NIT
Sbjct 355 EAAQKWKEVALASEEDYPSQIEAHWGQSVNKAYSDMCQWLGSINIDLSINEVVNNNITGE 414
Query 390 NEALIAGKGVGTGQSSESF-YAKDWGILMCIYHSVPLLDYVLSAPDPQLFTSENTSFPVP 448
N A IAGKG +G S +F +GI+MC++H +P LDY+ SAP + FP+P
Sbjct 415 NAADIAGKGTMSGNGSINFNVGGQYGIVMCVFHVLPQLDYITSAPHFGTTLTNVLDFPIP 474
Query 449 ELDSIGLEPISVAYYSNNPTELPSASGLLSDPTVTVGYLPRYFAWKTSLDYVLGAFTTTE 508
E D IG+E + V NP + P P + GY P+Y+ WKT+LD +G F +
Sbjct 475 EFDKIGMEQVPVI-RGLNPVK-PKDGDFKVSPNLYFGYAPQYYNWKTTLDKSMGEFRRSL 532
Query 509 KEWVAP 514
K W+ P
Sbjct 533 KTWIIP 538
>gi|575094321|emb|CDL65708.1| unnamed protein product [uncultured bacterium]
Length=642
Score = 246 bits (628), Expect = 2e-69, Method: Compositional matrix adjust.
Identities = 177/574 (31%), Positives = 279/574 (49%), Gaps = 64/574 (11%)
Query 2 SLFNLSSVKNHPRRSGFDLSSKVAFTAKVGELLPVKWTLTMPGDKFSLKEQHFTRTQPVN 61
++ L +KN P R+ FDLS + FTAKVGELLP PGD + +FTRT P+
Sbjct 6 NIMGLHGLKNKPSRNSFDLSHRNMFTAKVGELLPCFVQELNPGDSVKVSSSYFTRTAPLQ 65
Query 62 TSAYTRVREYYDWFWCPLHLLWRNAPEVISQIQQNV------QHASSFDGSVLLGSNMPC 115
++A+TR+RE +F+ P LW+ + + +N + ASS G+ + + MPC
Sbjct 66 SNAFTRLRENVQYFFVPYSALWKYFDSQVLNMTKNANGGDISRIASSLVGNQKVTTQMPC 125
Query 116 LSADQISQSLDQLKSKQNY-----------FGFDRADLAYKLLQYLRYGNVRTGVGS--- 161
++ + L + ++ G R + KLLQ L YGN +
Sbjct 126 VNYKTLHAYLLKFINRSTVGSDGSVGPEFNRGCYRHAESAKLLQLLGYGNFPEQFANFKV 185
Query 162 NGARNYGTSIDVKDGTYNQNRAYNHALSIFPILAYKKFCQDYFRLTQWQDSAPYLWNIDY 221
N ++ + + KD TYN N Y LSIF +LAY K C D++ QWQ L N+DY
Sbjct 186 NNDKHNQSGQNFKDVTYN-NSPY---LSIFRLLAYHKICNDHYLYRQWQPYNASLCNVDY 241
Query 222 YDGKG----AVTILPTSLTSSAAYFEGDTFFDLEYCNWNKDMFFGILPDAQFGDTSVVDI 277
++ S+ + E D+ + N D F G+LP +QFG SVV++
Sbjct 242 LTPNSSSLLSIDDALLSIPDDSIKAEKLNLLDMRFSNLPLDYFTGVLPTSQFGSESVVNL 301
Query 278 SYG-VTGAPVVT-------------------KQNLQSPTNSSVSI----GTDDANSKTL- 312
+ G +G+ V+ +Q + S N ++ + GT ++ T
Sbjct 302 NLGNASGSAVLNGTTSKDSGRWRTTTGEWEMEQRVASSANGNLKLDNSNGTFISHDHTFS 361
Query 313 --IASGTNLT--LDVLALRRGEALQRFREISLCTPLNYRSQIKAHFGVDVGAAMSGMSTY 368
+A T+L+ L ++ALR A Q+++EI L ++++SQ++AHFG+ S +
Sbjct 362 GNVAINTSLSGNLSIIALRNALAAQKYKEIQLANDVDFQSQVEAHFGIKPDEKNEN-SLF 420
Query 369 IGGEASSLDISEVVNTNITETNEALIAGKGVGTGQSSESFYAKDWGILMCIYHSVPLLDY 428
IGG +S ++I+E +N N++ N+A G G +S F AK +G+++ IY P+LD+
Sbjct 421 IGGSSSMININEQINQNLSGDNKATYGAAPQGNGSASIKFTAKTYGVVIGIYRCTPVLDF 480
Query 429 VLSAPDPQLFTSENTSFPVPELDSIGL------EPISVAYYSNNPTELPSASGLLSDPTV 482
D LF ++ + F +PE+DSIG+ E + A Y++ G D +
Sbjct 481 AHLGIDRTLFKTDASDFVIPEMDSIGMQQTFRCEVAAPAPYNDEFKAFRVGDGSSPDMSE 540
Query 483 TVGYLPRYFAWKTSLDYVLGAFTTTEKEWVAPIT 516
T GY PRY +KTS D GAF + K WV I
Sbjct 541 TYGYAPRYSEFKTSYDRYNGAFCHSLKSWVTGIN 574
>gi|494308783|ref|WP_007173938.1| hypothetical protein [Prevotella bergensis]
gi|270333035|gb|EFA43821.1| putative capsid protein (F protein) [Prevotella bergensis DSM
17361]
Length=553
Score = 177 bits (448), Expect = 5e-45, Method: Compositional matrix adjust.
Identities = 143/531 (27%), Positives = 240/531 (45%), Gaps = 73/531 (14%)
Query 1 MSLFNLSSVKNHPRRSGFDLSSKVAFTAKVGELLPVKWTLTMPGDKFSLKEQHFTRTQPV 60
+S+ + + + + R+ FDLS + FTA G LLPV +P D + Q F RT P+
Sbjct 3 VSIPKIKATRPNRNRNAFDLSQRHLFTAHAGMLLPVLNLDLIPHDHVEINAQDFMRTLPM 62
Query 61 NTSAYTRVREYYDWFWCPLHLLWRNAPEVISQIQQNVQHASSFDGSVLLGSN---MPCLS 117
NT+A+ +R Y++F+ P H LW + I+ + N H+S+ + S+ G++ +P +
Sbjct 63 NTAAFASMRGVYEFFFVPYHQLWAQFDQFITGM--NDFHSSA-NKSIQGGTSPLQVPYFN 119
Query 118 ADQISQSL-----------DQLKSKQNYFGFDRADLAYKLLQYLRYGNVRTGVGSNGARN 166
D + SL D L+ K Y A++LL L YG +
Sbjct 120 VDSVFNSLNTGKESGSGSTDDLQYKFKY-------GAFRLLDLLGYG--------RKFDS 164
Query 167 YGTSIDVKDGTYNQNRAYNHALSIFPILAYKKFCQDYFRLTQWQDSAPYLWNIDYYDGKG 226
+GT+ N YN S+F ILAY K QDY+R + +++ +N D + G
Sbjct 165 FGTAYPDNVSGLKNNLDYN--CSVFRILAYNKIYQDYYRNSNYENFDTDSFNFDKFKGGL 222
Query 227 AVTILPTSLTSSAAYFEGDTFFDLEYCNWNKDMFFGILPDAQFGDTSVVDISYGVTGAPV 286
+ L F L Y N D F + F T+ + + AP
Sbjct 223 VDAKVVADL------------FKLRYRNAQTDYFTNLRQSQLFSFTTAFEDVDNINIAPR 270
Query 287 -VTKQNLQSPTNSSVSIGTDDANSKTLIASGTNLTLDVLALRRGEALQRFREISLCTPLN 345
K + + T + + TD + ++S LR A+ + +++
Sbjct 271 DYVKSDGSNFTRVNFGVDTDSSEGDFSVSS----------LRAAFAVDKLLSVTMRAGKT 320
Query 346 YRSQIKAHFGVDVGAAMSGMSTYIGGEASSLDISEVVNTNITETNE--------ALIAGK 397
++ Q++AH+GV++ + G Y+GG S + +S+V T+ T E +AGK
Sbjct 321 FQDQMRAHYGVEIPDSRDGRVNYLGGFDSDMQVSDVTQTSGTTATEYKPEAGYLGRVAGK 380
Query 398 GVGTGQSSESFYAKDWGILMCIYHSVPLLDYVLSAPDPQLFTSENTSFPVPELDSIGLEP 457
G G+G+ F AK+ G+LMCIY VP + Y + DP + + + PE +++G++P
Sbjct 381 GTGSGRGRIVFDAKEHGVLMCIYSLVPQIQYDCTRLDPMVDKLDRFDYFTPEFENLGMQP 440
Query 458 ISVAYYSNNPTELPSASGLLSDPTVTVGYLPRYFAWKTSLDYVLGAFTTTE 508
++ +Y S+ T P +GY PRY +KT+LD G F ++
Sbjct 441 LNSSYISSFCTTDPK--------NPVLGYQPRYSEYKTALDVNHGQFAQSD 483
>gi|496521299|ref|WP_009229582.1| capsid protein [Prevotella sp. oral taxon 317]
gi|288330570|gb|EFC69154.1| putative capsid protein (F protein) [Prevotella sp. oral taxon
317 str. F0108]
Length=541
Score = 170 bits (430), Expect = 9e-43, Method: Compositional matrix adjust.
Identities = 151/528 (29%), Positives = 237/528 (45%), Gaps = 77/528 (15%)
Query 1 MSLFNLSSVK----NHPRRSGFDLSSKVAFTAKVGELLPVKWTLTMPGDKFSLKEQHFTR 56
MSL + +K N PR S FDLS K +TA G LLPV M D ++ Q F R
Sbjct 1 MSLKKVPQIKPSRANRPR-SAFDLSQKHLYTAPAGALLPVLSVDLMFHDHIRIQAQDFMR 59
Query 57 TQPVNTSAYTRVREYYDWFWCPLHLLWRNAPEVISQIQQ-NVQHASSFDGSVLLGSNMPC 115
T P+N++A+ +R Y++F+ P LW + I+ + SS G L S +P
Sbjct 60 TMPMNSAAFISMRGVYEFFFVPYSQLWHPYDQFITSMNDYRSSVVSSAAGDKALDS-VPN 118
Query 116 LSADQISQSLDQLKSKQNYFGFDRADLAYKLLQYLRYGNVRTGVGSNGARNYGTSIDVKD 175
+ + + + + ++ ++ FG+ ++ + +L+ L YG T + T + +
Sbjct 119 VKLADMYKFVRE-RTDKDIFGYPHSNNSCRLMDLLGYGKPIT--------SSKTPVPL-- 167
Query 176 GTYNQNRAYNHALSIFPILAYKKFCQDYFRLTQWQDSAPYLWNIDYYDGKGAVTILPTSL 235
Y +++F +LAY K DY+R T ++ Y +NID+ G T +PT+
Sbjct 168 -------LYTGNVNLFRLLAYNKIYSDYYRNTTYEGVDVYSFNIDHKKG----TFVPTAD 216
Query 236 TSSAAYFEGDTFFDLEYCNWNKDMFFGILPDAQF--GDTSVVDISYGVTGAPVVTKQNLQ 293
E + +L Y N D + + P F G S + L
Sbjct 217 -------EFKKYLNLHYRNAPLDFYTNLRPTPLFTIGSDSFSSV------------LQLS 257
Query 294 SPTNSSVSIGTDDANSKTLIASGTNLTLDVLALRRGEALQRFREISLCTPLNYRSQIKAH 353
PT S+ + D NS L + ++ L+V A+R AL + IS+ Y QI+AH
Sbjct 258 DPTGSAGF--SADGNSAKLNMASPDV-LNVSAIRSAFALDKLLSISMRAGKTYAEQIEAH 314
Query 354 FGVDVGAAMSGMSTYIGGEASSLDISEV------VNTNITETNEALIA-------GKGVG 400
FGV V G Y+GG S++ + +V N N++E A +A GKG G
Sbjct 315 FGVTVSEGRDGQVYYLGGFDSNVQVGDVTQTSGTTNPNVSEVGNAKLAGYLGKITGKGTG 374
Query 401 TGQSSESFYAKDWGILMCIYHSVPLLDYVLSAPDPQLFTSENTSFPVPELDSIGLEPISV 460
+G F AK+ G+LMCIY VP + Y DP + + +PE +++G++PI
Sbjct 375 SGYGEIQFDAKEPGVLMCIYSVVPAMQYDCMRLDPFVAKQTRGDYFIPEFENLGMQPIVP 434
Query 461 AYYSNNPTELPSASGLLSDPTVTVGYLPRYFAWKTSLDYVLGAFTTTE 508
A+ S N + S G+ PRY +KT+ D G F E
Sbjct 435 AFVSLNRAKDNS-----------YGWQPRYSEYKTAFDINHGQFANGE 471
>gi|517172762|ref|WP_018361580.1| hypothetical protein [Prevotella nanceiensis]
Length=568
Score = 158 bits (399), Expect = 1e-38, Method: Compositional matrix adjust.
Identities = 140/521 (27%), Positives = 235/521 (45%), Gaps = 66/521 (13%)
Query 15 RSGFDLSSKVAFTAKVGELLPVKWTLTMPGDKFSLKEQHFTRTQPVNTSAYTRVREYYDW 74
R+ FD+S + FTA G LLPV +P D + F RT P+N++A+ +R Y++
Sbjct 18 RNAFDISQRHLFTAPAGALLPVLSLDLLPHDHVEINASDFMRTLPMNSAAFMSMRGVYEF 77
Query 75 FWCPLHLLWRNAPEVISQIQQNVQHASSFDGSVLLGSNMPCLSADQISQSLDQLKSK--Q 132
++ P LW + I+ + + SSF + + C+S D + + +D K+ +
Sbjct 78 YFVPYKQLWSGFDQFITGMS---DYKSSFMYAFKGKTPPSCVSFD-VQKLVDWCKTNTAK 133
Query 133 NYFGFDRADLAYKLLQYLRYGNVRTGVGSNGARNYGTSIDVKDGTYNQNRAYNHALSIFP 192
+ GFD+ Y++L L YG G T++ + F
Sbjct 134 DIHGFDKNKGVYRILDLLGYGKYANSAGVPYTNPTSTTMG--------------KCTPFR 179
Query 193 ILAYKKFCQDYFRLTQWQDSAPYLWNIDYYDGKGAVTILPTSLTSSAAYFEGDTFFDLEY 252
LAY+K D++R T +++ +N+D + G G V ++ + ++ +F L Y
Sbjct 180 GLAYQKIYNDFYRNTTYEEYQLESFNVDMFYGSGKVK---ETIPNEPWDYD---WFTLRY 233
Query 253 CNWNKDMFFGILPDAQFGDTSVVDIS--YGVTGAPVVTKQNLQSPTNSSVSIGTDDANSK 310
N KD+ + P F S+ D + + G+ +V ++ +V+ GT +
Sbjct 234 RNAQKDLLTNVRPTPLF---SIDDFNPQFFTGGSDIVMEK------GPNVTGGTHEYRDS 284
Query 311 TLIASGTNLT----------LDVLALRRGEALQRFREISLCTPLNYRSQIKAHFGVDVGA 360
+I G NL + V +R AL++ +++ Y+ Q++AHFG+ V
Sbjct 285 VVIV-GKNLKENGVDSKRTMISVADIRNAFALEKLASVTMRAGKTYKEQMEAHFGISVEE 343
Query 361 AMSGMSTYIGGEASSLDISEVVN---TNITETNE-------ALIAGKGVGTGQSSESFYA 410
G TYIGG S++ + +V T +T T + GK G+G F A
Sbjct 344 GRDGRCTYIGGFDSNIQVGDVTQSSGTTVTGTKDTSFGGYLGRTTGKATGSGSGHIRFDA 403
Query 411 KDWGILMCIYHSVPLLDYVLSAPDPQLFTSENTSFPVPELDSIGLEPI---SVAYYSNNP 467
K+ GILMCIY VP + Y DP + E F VPE +++G++P+ +++Y NN
Sbjct 404 KEHGILMCIYSLVPDVQYDSKRVDPFVQKIERGDFFVPEFENLGMQPLFAKNISYKYNNN 463
Query 468 TELPSASGLLSDPTVTVGYLPRYFAWKTSLDYVLGAFTTTE 508
T L + G+ PRY +KT+LD G F E
Sbjct 464 TANSRIKNLGA-----FGWQPRYSEYKTALDINHGQFVHQE 499
>gi|494306153|ref|WP_007173049.1| hypothetical protein [Prevotella bergensis]
gi|270333881|gb|EFA44667.1| putative capsid protein (F protein) [Prevotella bergensis DSM
17361]
Length=519
Score = 154 bits (388), Expect = 3e-37, Method: Compositional matrix adjust.
Identities = 132/506 (26%), Positives = 225/506 (44%), Gaps = 88/506 (17%)
Query 33 LLPVKWTLTMPGDKFSLKEQHFTRTQPVNTSAYTRVREYYDWFWCPLHLLWRNAPEVISQ 92
LLPV +P D + Q F RT P+NT+A+ +R Y++F+ P H LW + I+
Sbjct 2 LLPVLNLDLIPHDHVEINAQDFMRTLPMNTAAFASMRGVYEFFFVPYHQLWAQFDQFITG 61
Query 93 IQQNVQHASSFDGSVLLGSN---MPCLSADQISQSL---DQLKSKQNYFGFDRADLAYKL 146
+ N H+S+ + S+ G++ +P + + + +++ D S Q+ + A++L
Sbjct 62 M--NDFHSSA-NKSIQGGTSPLQVPYFNLESVFKNIIERDSTPSFQDDLQYRFKYGAFRL 118
Query 147 LQYLRYGNVRTGVGSNGARNYGTSIDVKDGTYNQNRAYNHALSIFPILAYKKFCQDYFRL 206
L L YG ++GT+ N YN S+F +LAY K QDY+R
Sbjct 119 LDLLGYGR--------KFDSFGTAYPDNVSGLKNNLDYN--CSVFRVLAYNKIYQDYYRN 168
Query 207 TQWQDSAPYLWNIDYYDG----------------KGAVTILPTSLTSSAAYFEGDTFFDL 250
+ +++ +N D + G + A T T+L S + F D
Sbjct 169 SNYENFDTDSFNFDKFKGGLVDAKVVADLFKLRYRNAQTDYFTNLRQSQLFTFIPEFSDD 228
Query 251 EYCNWNKDMFFGILPDAQFGDTSVVDISYGVTGAPVVTKQNLQSPTNSSVSIGTDDANSK 310
E+ N+++D Q+ D S + + L P + ++G
Sbjct 229 EHLNFDRD---------QYADQSKSNFT------------QLNFPVDVDNNLGY------ 261
Query 311 TLIASGTNLTLDVLALRRGEALQRFREISLCTPLNYRSQIKAHFGVDVGAAMSGMSTYIG 370
V +LR A+ + +++ ++ Q++AH+GV++ + G Y+G
Sbjct 262 ----------FSVSSLRSAFAVDKLLSVTMRAGKTFQDQMRAHYGVEIPDSRDGRVNYLG 311
Query 371 GEASSLDISEVVNTNITETNE--------ALIAGKGVGTGQSSESFYAKDWGILMCIYHS 422
G S L +S+V T+ T E IAGKG G+G+ F AK+ G+LMCIY
Sbjct 312 GFDSDLQVSDVTQTSGTTATEYKPEAGYLGRIAGKGTGSGRGRIVFDAKEHGVLMCIYSL 371
Query 423 VPLLDYVLSAPDPQLFTSENTSFPVPELDSIGLEPISVAYYSNNPTELPSASGLLSDPTV 482
VP + Y + DP + + F PE +++G++P++ +Y S+ T P
Sbjct 372 VPQIQYDCTRLDPMVDKLDRFDFFTPEFENLGMQPLNSSYISSFCTPDPK--------NP 423
Query 483 TVGYLPRYFAWKTSLDYVLGAFTTTE 508
+GY PRY +KT+LD G F +
Sbjct 424 VLGYQPRYSEYKTALDINHGQFAQND 449
Lambda K H a alpha
0.318 0.133 0.407 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 3834607117410