bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: All non-redundant GenBank CDS translations+PDB+SwissProt+PIR+PRF
excluding environmental samples from WGS projects
49,011,213 sequences; 17,563,301,199 total letters
Query= Contig-31_CDS_annotation_glimmer3.pl_2_5
Length=290
Score E
Sequences producing significant alignments: (Bits) Value
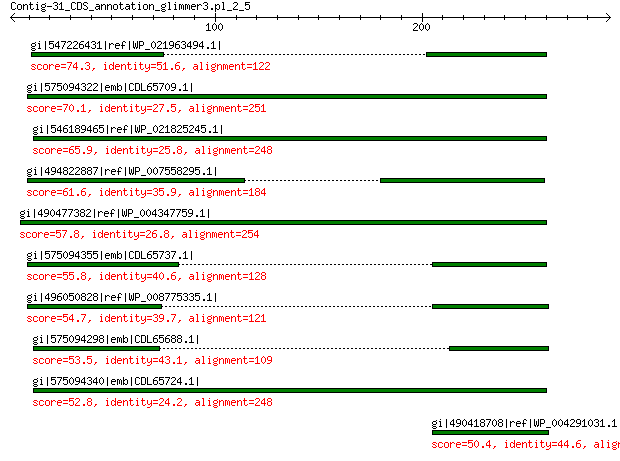
gi|547226431|ref|WP_021963494.1| predicted protein 74.3 8e-12
gi|575094322|emb|CDL65709.1| unnamed protein product 70.1 3e-10
gi|546189465|ref|WP_021825245.1| hypothetical protein 65.9 8e-09
gi|494822887|ref|WP_007558295.1| hypothetical protein 61.6 2e-07
gi|490477382|ref|WP_004347759.1| hypothetical protein 57.8 4e-06
gi|575094355|emb|CDL65737.1| unnamed protein product 55.8 1e-05
gi|496050828|ref|WP_008775335.1| hypothetical protein 54.7 4e-05
gi|575094298|emb|CDL65688.1| unnamed protein product 53.5 7e-05
gi|575094340|emb|CDL65724.1| unnamed protein product 52.8 1e-04
gi|490418708|ref|WP_004291031.1| hypothetical protein 50.4 7e-04
>gi|547226431|ref|WP_021963494.1| predicted protein [Prevotella sp. CAG:1185]
gi|524103383|emb|CCY83995.1| predicted protein [Prevotella sp. CAG:1185]
Length=498
Score = 74.3 bits (181), Expect = 8e-12, Method: Compositional matrix adjust.
Identities = 31/64 (48%), Positives = 47/64 (73%), Gaps = 0/64 (0%)
Query 11 KCENPRIIQNKYTGDFVKVDCGECPYCLIKKSDRATQKCDFVKFNHKYCYFVTLTYNSEY 70
KC +PR +QNKYTG+ ++V CG C CL +++D+ + C + +HKYC F TLTY+++Y
Sbjct 10 KCYHPRHVQNKYTGEVIQVGCGVCKACLKRRADKMSFLCAIEEQSHKYCMFATLTYSNDY 69
Query 71 VPKM 74
VP+M
Sbjct 70 VPRM 73
Score = 61.2 bits (147), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 32/58 (55%), Positives = 38/58 (66%), Gaps = 2/58 (3%)
Query 202 QFKGLLKYINIRDYQLFSKRLRKYLSKKIGKYEKIHSYVVSEYSPKTLRPHFHILFFF 259
G L Y + RD QLF KR+RK LSK EKI Y+VSEY PKT R H+H+LFF+
Sbjct 125 NLDGYLSYTSKRDAQLFLKRVRKNLSKYSD--EKIRYYIVSEYGPKTFRAHYHVLFFY 180
>gi|575094322|emb|CDL65709.1| unnamed protein product [uncultured bacterium]
Length=499
Score = 70.1 bits (170), Expect = 3e-10, Method: Compositional matrix adjust.
Identities = 69/251 (27%), Positives = 107/251 (43%), Gaps = 66/251 (26%)
Query 9 FNKCENPRIIQNKYTGDFVKVDCGECPYCLIKKSDRATQKCDFVKFNHKYCYFVTLTYNS 68
F KC +P ++++ G +V CG+C C K + K ++ KYCYF+TLTY+
Sbjct 5 FVKCFSPLVLRDP-RGYPYQVPCGKCIACHNNKRSSLSLKLRLEEYTSKYCYFLTLTYDD 63
Query 69 EYVPKMSLTQIDDYLTEWLPVRPPKSIGTQLVARMVMDSRVNKKIPNFVSAKVNRPYMLE 128
+ +P S+ +D TE++ + PY
Sbjct 64 DNLPLFSVG-LDTCATEFVRIY---------------------------------PY--- 86
Query 129 HLHFIEAERYKALSLRYPNFGSKFRPYILRSILRKSPLQRFKDEYFEELVWMLPELAESL 188
+ER LR +F S F S L F +++ +++ + +
Sbjct 87 ------SER-----LRNDSFISDF----------CSDLHNFDNDFVDKMDYYSDYVINYE 125
Query 189 KKKNNTDANGAFPQFKGLLKYINIRDYQLFSKRLRKYLSKKIGKYEKIHSYVVSEYSPKT 248
K + + G GL + RD QLF KRLRK++ K G EKI Y++ EY K+
Sbjct 126 SKYHKSCVYG-----HGLYALLYYRDIQLFLKRLRKHIYKYYG--EKIRFYIIGEYGTKS 178
Query 249 LRPHFHILFFF 259
LRPH+H L FF
Sbjct 179 LRPHWHCLLFF 189
>gi|546189465|ref|WP_021825245.1| hypothetical protein [Prevotella salivae]
gi|544001993|gb|ERK01417.1| hypothetical protein HMPREF9145_2741 [Prevotella salivae F0493]
Length=586
Score = 65.9 bits (159), Expect = 8e-09, Method: Compositional matrix adjust.
Identities = 64/256 (25%), Positives = 116/256 (45%), Gaps = 39/256 (15%)
Query 12 CENPRIIQNKYTGDFVKVDCGECPYCLIKKSDRATQKCDFVKFNHKYCYFVTLTYNSEYV 71
C +P I++N G V CG+C CL KK+ + ++K+C F TLTY++E+V
Sbjct 17 CTSPIIVEN--NGRKYAVACGKCECCLHKKATIWRTRLRQEMKDNKFCLFFTLTYDNEHV 74
Query 72 PKMSLTQIDDYLT----EWLPVRPPKSIGTQLVARMVMDSRVNKKIPNFVSAKVNRPYML 127
P + +D+ T E + ++ ++ T R V S +P + V + +
Sbjct 75 PFFGRAKNNDFYTLDGEEGVQLKGDDNL-TYSPKRSVPTSLCRDGVPTITNFDVCDSFAV 133
Query 128 EHLHFIEAERYKALSLRYPNFGSKFRPYILRSILRKSPLQRFKDEYFEELVWMLPELAES 187
+ ++A++ F +FR ++ +++ L F+D+ F ++
Sbjct 134 --VSRVDAQK----------FMKRFRWHLFHLLVKHYKLI-FQDKLFTFTQYL------- 173
Query 188 LKKKNNTDANGAFPQFKGLLKYINIRDYQLFSKRLRKYLS----KKIGKYEKIHSYVVSE 243
+G+ P FK L ++ Y L+ + YL+ KK + + ++ SE
Sbjct 174 -------GYDGSIP-FKEWLDDLDTETYDLYYSVYQYYLTDYEKKKESCKQSVRYFICSE 225
Query 244 YSPKTLRPHFHILFFF 259
Y+P T RPHFH LF+F
Sbjct 226 YTPTTFRPHFHGLFWF 241
>gi|494822887|ref|WP_007558295.1| hypothetical protein [Bacteroides plebeius]
gi|198272100|gb|EDY96369.1| hypothetical protein BACPLE_00805 [Bacteroides plebeius DSM 17135]
Length=545
Score = 61.6 bits (148), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 35/79 (44%), Positives = 45/79 (57%), Gaps = 7/79 (9%)
Query 180 MLPELAESLKKKNNTDANGAFPQFKGLLKYINIRDYQLFSKRLRKYLSKKIGKYEKIHSY 239
M P+L +K+ N N +KG Y++ R+ QLF KRLRKYL K G +KI +
Sbjct 96 MTPQLMNEYQKRVNYRIN-----YKGRFPYLSKRELQLFMKRLRKYLDKYEG--QKIRFF 148
Query 240 VVSEYSPKTLRPHFHILFF 258
EY P + RPHFHIL F
Sbjct 149 ATGEYGPLSFRPHFHILLF 167
Score = 52.4 bits (124), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 31/108 (29%), Positives = 53/108 (49%), Gaps = 3/108 (3%)
Query 9 FNKCENPRIIQNKYTGDFVKVDCGECPYCLIKKSDRATQKCDFVKFNHKYCYFVTLTYNS 68
F C P+ I+NKYTG+ + V C C C ++ + + CDF K F+TLT++
Sbjct 5 FVSCLEPQRIKNKYTGEEMVVACKHCVACEQLRNFKYSNLCDFESLTAKKTVFLTLTFDD 64
Query 69 EYVPKMSLTQIDDYLTEWLPVRPPKSIGTQLVARMVMDS---RVNKKI 113
++VP+ ++ D + +G L+ +M+ RVN +I
Sbjct 65 KFVPQFRFYKVGDDEYIMRDADTGEYLGRTLMTPQLMNEYQKRVNYRI 112
>gi|490477382|ref|WP_004347759.1| hypothetical protein [Prevotella buccalis]
gi|281300711|gb|EFA93042.1| hypothetical protein HMPREF0650_1078 [Prevotella buccalis ATCC
35310]
Length=582
Score = 57.8 bits (138), Expect = 4e-06, Method: Compositional matrix adjust.
Identities = 68/257 (26%), Positives = 102/257 (40%), Gaps = 73/257 (28%)
Query 6 FKYFNKCENPRIIQNKYTGDFVKVDCGECPYCLIKKSDRATQKCDFVKFNHKYCYFVTLT 65
K C NPR + N ++ C +C CL +K+ + + HKY F TLT
Sbjct 7 LKIVGNCLNPRKVYNPSLHGWMYCSCDKCTACLNQKATTLSNRARAEIEQHKYSVFFTLT 66
Query 66 YNSEYVPKMSLTQIDDYLTEWLPVRPPKSIGTQLVARMVMDSRVNKKIPNFVSAKVNRPY 125
Y++E++PK + Q + E + RP + R+V D + + N S +N+
Sbjct 67 YDNEHLPKYEVFQDSN---EVIQYRP--------IGRLV-DDSSSDMLSN--SCPINKYN 112
Query 126 MLEHLHFIEAERYKALSLRYPNFGSKFRPYILRSILRKSPLQRFKDEYFEELVWMLPELA 185
E+L+ + S F P P++ ++D Y
Sbjct 113 NYENLYQFDE--------------STFIP----------PIENYEDIY------------ 136
Query 186 ESLKKKNNTDANGAFPQFKGLLKYINIRDYQLFSKRLRKYLSK--KIGKYE-KIHSYVVS 242
F + K +D Q F KRLR +SK I K E KI Y+ S
Sbjct 137 ----------------HFGVVCK----KDIQNFLKRLRWRISKIPNITKDESKIRYYISS 176
Query 243 EYSPKTLRPHFHILFFF 259
EY P T RPH+H + FF
Sbjct 177 EYGPTTYRPHYHGILFF 193
>gi|575094355|emb|CDL65737.1| unnamed protein product [uncultured bacterium]
Length=517
Score = 55.8 bits (133), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 26/73 (36%), Positives = 40/73 (55%), Gaps = 0/73 (0%)
Query 9 FNKCENPRIIQNKYTGDFVKVDCGECPYCLIKKSDRATQKCDFVKFNHKYCYFVTLTYNS 68
F +C P+ + N Y D++ V CG+C C K+ R + HK+C F TLTY +
Sbjct 8 FIRCLEPKRVFNPYLNDWLLVPCGKCRACQCSKASRYKLQIQLEASQHKFCIFGTLTYAN 67
Query 69 EYVPKMSLTQIDD 81
Y+P++SL +D
Sbjct 68 TYIPRLSLVPYND 80
Score = 48.1 bits (113), Expect = 0.005, Method: Compositional matrix adjust.
Identities = 26/55 (47%), Positives = 32/55 (58%), Gaps = 2/55 (4%)
Query 205 GLLKYINIRDYQLFSKRLRKYLSKKIGKYEKIHSYVVSEYSPKTLRPHFHILFFF 259
G + Y+ RD QLF KRLRK LSK K+ + + EY P RPH+H L FF
Sbjct 121 GDVPYLRKRDLQLFIKRLRKNLSKYSDA--KVRYFAMGEYGPVHFRPHYHFLLFF 173
>gi|496050828|ref|WP_008775335.1| hypothetical protein [Bacteroides sp. 2_2_4]
gi|229448892|gb|EEO54683.1| hypothetical protein BSCG_01608 [Bacteroides sp. 2_2_4]
Length=497
Score = 54.7 bits (130), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 24/65 (37%), Positives = 38/65 (58%), Gaps = 0/65 (0%)
Query 9 FNKCENPRIIQNKYTGDFVKVDCGECPYCLIKKSDRATQKCDFVKFNHKYCYFVTLTYNS 68
F KC +P+ I N YT + + V CG C C + K+ R +CD + K+ F+TLTY +
Sbjct 6 FCKCLHPKRIMNPYTKESMVVPCGHCQACTLAKNSRYAFQCDLESYTAKHTLFITLTYAN 65
Query 69 EYVPK 73
++P+
Sbjct 66 RFIPR 70
Score = 47.4 bits (111), Expect = 0.007, Method: Compositional matrix adjust.
Identities = 24/56 (43%), Positives = 34/56 (61%), Gaps = 1/56 (2%)
Query 205 GLLKYINIRDYQLFSKRLRKYLSKKIGKYEKIHSYVVSEYSPKTLRPHFHILFFFR 260
G + Y+ D QLF KRLR Y++K+ EK+ + V EY P RPH+H+L F +
Sbjct 115 GDVPYLRKTDLQLFLKRLRYYVTKQKPS-EKVRYFAVGEYGPVHFRPHYHLLLFLQ 169
>gi|575094298|emb|CDL65688.1| unnamed protein product [uncultured bacterium]
Length=478
Score = 53.5 bits (127), Expect = 7e-05, Method: Compositional matrix adjust.
Identities = 25/61 (41%), Positives = 39/61 (64%), Gaps = 0/61 (0%)
Query 12 CENPRIIQNKYTGDFVKVDCGECPYCLIKKSDRATQKCDFVKFNHKYCYFVTLTYNSEYV 71
C N R I+NKYTG + V CG+CP CL +K++ + K + + C+FVTL Y++ ++
Sbjct 2 CINKREIRNKYTGQKLYVSCGKCPACLQEKANASAYKIRNNQSSELSCFFVTLNYDNNHI 61
Query 72 P 72
P
Sbjct 62 P 62
Score = 43.9 bits (102), Expect = 0.11, Method: Compositional matrix adjust.
Identities = 22/48 (46%), Positives = 28/48 (58%), Gaps = 0/48 (0%)
Query 213 RDYQLFSKRLRKYLSKKIGKYEKIHSYVVSEYSPKTLRPHFHILFFFR 260
+D QLF KRLR+ L +K G I + SEY P T R HFH+ F +
Sbjct 152 KDIQLFFKRLRQSLYRKFGFRPFIQYFQTSEYGPTTYRAHFHLCIFVK 199
>gi|575094340|emb|CDL65724.1| unnamed protein product [uncultured bacterium]
Length=486
Score = 52.8 bits (125), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 60/250 (24%), Positives = 97/250 (39%), Gaps = 54/250 (22%)
Query 12 CENPRIIQNKYTGDFVKVDCGECPYCLIKKSDRATQKCDFVKFNHKYCY--FVTLTYNSE 69
C N + NKY G VDCG CP CL +K++++ K ++ Y + FVTLTY++
Sbjct 6 CTNRIKVTNKYVGRSFYVDCGHCPSCLQRKANKSCCKI-INEYGRPYSFMCFVTLTYDN- 63
Query 70 YVPKMSLTQIDDYLTEWLPVRPPKSIGTQLVARMVMDSRVNKKIPNFVSAKVNRPYMLEH 129
E +P P + + L V + Y + H
Sbjct 64 ---------------EHIPYIHPDTDYSHLY--------------------VGKSYYVRH 88
Query 130 LHFIEAERYKALSLRYPNFGSKFRPYILRSILRKSPLQRFKDEYFEELVWMLPELAESLK 189
+ + + L L G I L + P + F++ Y ++ + +
Sbjct 89 SRIFDKDGVENLPLGVYRNGK----LIDTVFLPEMPKEVFRN-YLCNTTGIVTKSRNGVV 143
Query 190 KKNNTDANGAFPQFKGLLKYINIRDYQLFSKRLRKYLSKKIGKYEKIHSYVVSEYSPKTL 249
+ + + G +D+ F KRLR L++ KI + SEY P T
Sbjct 144 LERDDNKVGILYD----------KDFVNFVKRLRINLTRNYNYEGKITYFKCSEYGPTTN 193
Query 250 RPHFHILFFF 259
RPHFH +F+F
Sbjct 194 RPHFHGIFWF 203
>gi|490418708|ref|WP_004291031.1| hypothetical protein [Bacteroides eggerthii]
gi|217986635|gb|EEC52969.1| hypothetical protein BACEGG_02720 [Bacteroides eggerthii DSM
20697]
Length=422
Score = 50.4 bits (119), Expect = 7e-04, Method: Compositional matrix adjust.
Identities = 25/56 (45%), Positives = 34/56 (61%), Gaps = 1/56 (2%)
Query 205 GLLKYINIRDYQLFSKRLRKYLSKKIGKYEKIHSYVVSEYSPKTLRPHFHILFFFR 260
G L Y+ D QLF KR R Y++K+ K EK+ + + EY P RPH+HIL F +
Sbjct 39 GYLPYLRKFDLQLFFKRFRYYVAKRFPK-EKVRYFAIGEYGPVHFRPHYHILLFLQ 93
Lambda K H a alpha
0.324 0.139 0.433 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 1498703630544