bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: All non-redundant GenBank CDS translations+PDB+SwissProt+PIR+PRF
excluding environmental samples from WGS projects
49,011,213 sequences; 17,563,301,199 total letters
Query= Contig-31_CDS_annotation_glimmer3.pl_2_7
Length=101
Score E
Sequences producing significant alignments: (Bits) Value
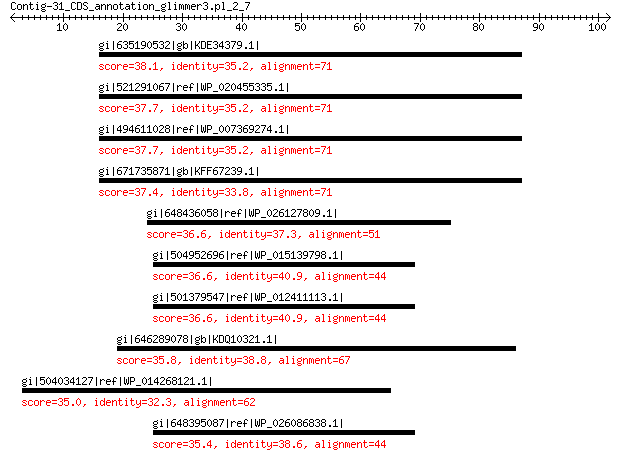
gi|635190532|gb|KDE34379.1| capsid protein 38.1 0.51
gi|521291067|ref|WP_020455335.1| oxidoreductase 37.7 0.54
gi|494611028|ref|WP_007369274.1| capsid protein 37.7 0.54
gi|671735871|gb|KFF67239.1| capsid protein 37.4 0.74
gi|648436058|ref|WP_026127809.1| peptide ABC transporter ATPase 36.6 1.4
gi|504952696|ref|WP_015139798.1| WD40 repeat-containing protein 36.6 1.7
gi|501379547|ref|WP_012411113.1| hypothetical protein 36.6 2.0
gi|646289078|gb|KDQ10321.1| hypothetical protein BOTBODRAFT_147832 35.8 3.2
gi|504034127|ref|WP_014268121.1| N-acetylmuramoyl-L-alanine amidase 35.0 4.8
gi|648395087|ref|WP_026086838.1| hypothetical protein 35.4 4.9
>gi|635190532|gb|KDE34379.1| capsid protein [Kosakonia radicincitans UMEnt01/12]
Length=385
Score = 38.1 bits (87), Expect = 0.51, Method: Compositional matrix adjust.
Identities = 25/76 (33%), Positives = 40/76 (53%), Gaps = 6/76 (8%)
Query 16 MGNQWQ---NQLTGELLNSKRWDNEMQAWREAINSANALLRGSSDAMNTFNTYENG--RY 70
M N++Q QL GEL+ S++ ++QA R AN++L + D ++ + YE+G R+
Sbjct 248 MINKFQANSEQLAGELIISQKTIGQLQAVRAPFFPANSILITTLDNLSVY-LYEDGHRRH 306
Query 71 ERNRPYTSDYEEYEQF 86
P E YEQ
Sbjct 307 IIENPKLDQVENYEQV 322
>gi|521291067|ref|WP_020455335.1| oxidoreductase [Enterobacter sp. R4-368]
gi|512647996|ref|YP_008108190.1| oxidoreductase [Enterobacter sp. R4-368]
gi|511526112|gb|AGN83652.1| oxidoreductase [Enterobacter sp. R4-368]
Length=390
Score = 37.7 bits (86), Expect = 0.54, Method: Compositional matrix adjust.
Identities = 25/76 (33%), Positives = 40/76 (53%), Gaps = 6/76 (8%)
Query 16 MGNQWQ---NQLTGELLNSKRWDNEMQAWREAINSANALLRGSSDAMNTFNTYENG--RY 70
M N++Q QL GEL+ S++ ++QA R AN++L + D ++ + YE+G R+
Sbjct 248 MINKFQPNTEQLAGELIISQKTIGQLQAVRAPFFPANSILITTLDNLSIY-LYEDGHRRH 306
Query 71 ERNRPYTSDYEEYEQF 86
P E YEQ
Sbjct 307 IIENPKLDQVENYEQV 322
>gi|494611028|ref|WP_007369274.1| capsid protein [Kosakonia radicincitans]
gi|396095568|gb|EJI93111.1| phage major capsid protein, P2 family [Kosakonia radicincitans
DSM 16656]
Length=390
Score = 37.7 bits (86), Expect = 0.54, Method: Compositional matrix adjust.
Identities = 25/76 (33%), Positives = 40/76 (53%), Gaps = 6/76 (8%)
Query 16 MGNQWQ---NQLTGELLNSKRWDNEMQAWREAINSANALLRGSSDAMNTFNTYENG--RY 70
M N++Q QL GEL+ S++ ++QA R AN++L + D ++ + YE+G R+
Sbjct 248 MINKFQPNTEQLAGELIISQKTIGQLQAVRAPFFPANSILITTLDNLSVY-LYEDGHRRH 306
Query 71 ERNRPYTSDYEEYEQF 86
P E YEQ
Sbjct 307 IIENPKLDQVENYEQV 322
>gi|671735871|gb|KFF67239.1| capsid protein [Pectobacterium carotovorum subsp. brasiliense]
Length=394
Score = 37.4 bits (85), Expect = 0.74, Method: Compositional matrix adjust.
Identities = 24/76 (32%), Positives = 41/76 (54%), Gaps = 6/76 (8%)
Query 16 MGNQWQN---QLTGELLNSKRWDNEMQAWREAINSANALLRGSSDAMNTFNTYENG--RY 70
+ N++Q+ QL GEL+ S++ ++QA R ANA++ + D ++ + YE+G R+
Sbjct 247 LINKFQDNSEQLAGELIISQKTIGQLQAVRAPFFPANAIMITTLDNLSIY-LYEDGHRRH 305
Query 71 ERNRPYTSDYEEYEQF 86
P E YEQ
Sbjct 306 IVENPKLDQVENYEQV 321
>gi|648436058|ref|WP_026127809.1| peptide ABC transporter ATPase [Nocardiopsis lucentensis]
Length=357
Score = 36.6 bits (83), Expect = 1.4, Method: Composition-based stats.
Identities = 19/51 (37%), Positives = 26/51 (51%), Gaps = 0/51 (0%)
Query 24 LTGELLNSKRWDNEMQAWREAINSANALLRGSSDAMNTFNTYENGRYERNR 74
L E L RW+ ++ A+NS N +LR +TF +ENGR R R
Sbjct 96 LGKEELRRFRWNEAAMVFQGAMNSFNPVLRIEDHFTDTFRAHENGRGRRPR 146
>gi|504952696|ref|WP_015139798.1| WD40 repeat-containing protein [Nostoc sp. PCC 7524]
gi|427730731|ref|YP_007076968.1| WD40 repeat-containing protein [Nostoc sp. PCC 7524]
gi|427366650|gb|AFY49371.1| WD40 repeat-containing protein [Nostoc sp. PCC 7524]
Length=1661
Score = 36.6 bits (83), Expect = 1.7, Method: Composition-based stats.
Identities = 18/44 (41%), Positives = 24/44 (55%), Gaps = 0/44 (0%)
Query 25 TGELLNSKRWDNEMQAWREAINSANALLRGSSDAMNTFNTYENG 68
TG+LL S WDN ++ WR LL+G SD++N NG
Sbjct 1522 TGQLLASAGWDNTVRLWRRDGTLLQTLLKGYSDSVNGVTFSPNG 1565
>gi|501379547|ref|WP_012411113.1| hypothetical protein [Nostoc punctiforme]
gi|186684904|ref|YP_001868100.1| hypothetical protein Npun_F4808 [Nostoc punctiforme PCC 73102]
gi|186467356|gb|ACC83157.1| WD-40 repeat protein [Nostoc punctiforme PCC 73102]
Length=1683
Score = 36.6 bits (83), Expect = 2.0, Method: Composition-based stats.
Identities = 18/44 (41%), Positives = 23/44 (52%), Gaps = 0/44 (0%)
Query 25 TGELLNSKRWDNEMQAWREAINSANALLRGSSDAMNTFNTYENG 68
T ELL S WDN ++ WR LL+G SD++N NG
Sbjct 1542 TDELLASASWDNTVKLWRRDGTLLKTLLKGYSDSVNAVTFSPNG 1585
>gi|646289078|gb|KDQ10321.1| hypothetical protein BOTBODRAFT_147832 [Botryobasidium botryosum
FD-172 SS1]
Length=958
Score = 35.8 bits (81), Expect = 3.2, Method: Composition-based stats.
Identities = 26/78 (33%), Positives = 40/78 (51%), Gaps = 15/78 (19%)
Query 19 QWQNQLTGELLNSKRWDNEMQAWREA-------INSANALLRGS----SDAMNTFNTYEN 67
QW N+L E L ++R E +W++A +N ANA+L+ DA N+ +N
Sbjct 669 QWVNELQ-EKLRARR--KEKASWQQAAVELRGEVNDANAMLKAQKKSLEDAQNSVFVLKN 725
Query 68 GRYERNRPYTSDYEEYEQ 85
E N P + YEEY++
Sbjct 726 -TVEENAPKVAKYEEYKR 742
>gi|504034127|ref|WP_014268121.1| N-acetylmuramoyl-L-alanine amidase [Serratia symbiotica]
gi|374313851|ref|YP_005060280.1| N-acetylmuramoyl-l-alanine amidase II [Serratia symbiotica str.
'Cinara cedri']
gi|363988077|gb|AEW44268.1| N-acetylmuramoyl-l-alanine amidase II [Serratia symbiotica str.
'Cinara cedri']
Length=448
Score = 35.0 bits (79), Expect = 4.8, Method: Composition-based stats.
Identities = 20/62 (32%), Positives = 31/62 (50%), Gaps = 0/62 (0%)
Query 3 RGKNVKQDAASDFMGNQWQNQLTGELLNSKRWDNEMQAWREAINSANALLRGSSDAMNTF 62
RG NV +D N+ N + +L+++R D+EM AW E + LL G+ D +
Sbjct 266 RGANVLVSIHADAAPNRSANGASVWVLSNRRADSEMAAWLEQYDKQPELLGGAGDMLANS 325
Query 63 NT 64
T
Sbjct 326 QT 327
>gi|648395087|ref|WP_026086838.1| hypothetical protein [Fischerella muscicola]
Length=1684
Score = 35.4 bits (80), Expect = 4.9, Method: Compositional matrix adjust.
Identities = 17/44 (39%), Positives = 23/44 (52%), Gaps = 0/44 (0%)
Query 25 TGELLNSKRWDNEMQAWREAINSANALLRGSSDAMNTFNTYENG 68
T LL S WDN ++ WR LL+G SD++N+ NG
Sbjct 1546 TDHLLASASWDNTVRLWRSDGTLLKTLLKGYSDSVNSVTFSPNG 1589
Lambda K H a alpha
0.311 0.126 0.378 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 438108004959