bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: All non-redundant GenBank CDS translations+PDB+SwissProt+PIR+PRF
excluding environmental samples from WGS projects
49,011,213 sequences; 17,563,301,199 total letters
Query= Contig-32_CDS_annotation_glimmer3.pl_2_1
Length=60
Score E
Sequences producing significant alignments: (Bits) Value
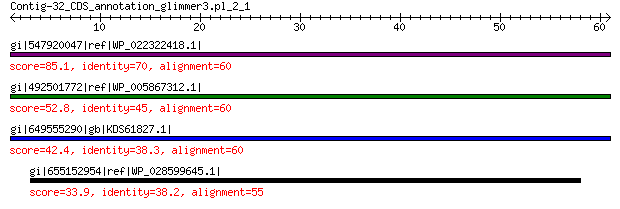
gi|547920047|ref|WP_022322418.1| putative uncharacterized protein 85.1 2e-18
gi|492501772|ref|WP_005867312.1| hypothetical protein 52.8 2e-06
gi|649555290|gb|KDS61827.1| hypothetical protein M095_3809 42.4 0.006
gi|655152954|ref|WP_028599645.1| peptidase M32 33.9 6.4
>gi|547920047|ref|WP_022322418.1| putative uncharacterized protein [Parabacteroides merdae CAG:48]
gi|524592959|emb|CDD13571.1| putative uncharacterized protein [Parabacteroides merdae CAG:48]
Length=259
Score = 85.1 bits (209), Expect = 2e-18, Method: Compositional matrix adjust.
Identities = 42/60 (70%), Positives = 50/60 (83%), Gaps = 1/60 (2%)
Query 1 MEALRKLRQDNDINAFRYSMERVFGNSSDVKDVASELVKRMGLLLMRPDIKELDQMFNPK 60
ME +RKL+QDNDINAFR MER+FG+SSD K+VASEL KRM + + R D +ELDQMFNPK
Sbjct 201 MEGIRKLQQDNDINAFRNRMERLFGDSSDAKNVASELFKRMMMYMFR-DSRELDQMFNPK 259
>gi|492501772|ref|WP_005867312.1| hypothetical protein [Parabacteroides distasonis]
gi|409230405|gb|EKN23269.1| hypothetical protein HMPREF1059_03254 [Parabacteroides distasonis
CL09T03C24]
Length=288
Score = 52.8 bits (125), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 27/60 (45%), Positives = 44/60 (73%), Gaps = 7/60 (12%)
Query 1 MEALRKLRQDNDINAFRYSMERVFGNSSDVKDVASELVKRMGLLLMRPDIKELDQMFNPK 60
++++R+L+QDNDINAFR MERV G SS A+++++R+ ++ +RP D++FNPK
Sbjct 236 LQSVRQLQQDNDINAFRNEMERVTGKSS----FATDMLRRL-IMALRPSAN--DRLFNPK 288
>gi|649555290|gb|KDS61827.1| hypothetical protein M095_3809 [Parabacteroides distasonis str.
3999B T(B) 4]
gi|649557306|gb|KDS63785.1| hypothetical protein M095_3404 [Parabacteroides distasonis str.
3999B T(B) 4]
gi|649559158|gb|KDS65545.1| hypothetical protein M096_4689 [Parabacteroides distasonis str.
3999B T(B) 6]
gi|649560567|gb|KDS66875.1| hypothetical protein M095_2448 [Parabacteroides distasonis str.
3999B T(B) 4]
gi|649561016|gb|KDS67303.1| hypothetical protein M095_2410 [Parabacteroides distasonis str.
3999B T(B) 4]
gi|649562727|gb|KDS68911.1| hypothetical protein M096_3341 [Parabacteroides distasonis str.
3999B T(B) 6]
Length=288
Score = 42.4 bits (98), Expect = 0.006, Method: Compositional matrix adjust.
Identities = 23/60 (38%), Positives = 39/60 (65%), Gaps = 7/60 (12%)
Query 1 MEALRKLRQDNDINAFRYSMERVFGNSSDVKDVASELVKRMGLLLMRPDIKELDQMFNPK 60
++++R+L QDNDINAFRY +E++ G K + + R L ++ P+ + D++FNPK
Sbjct 236 LQSVRQLTQDNDINAFRYRIEKITG-----KGPLLDDLIRRLLTILAPN--DFDRLFNPK 288
>gi|655152954|ref|WP_028599645.1| peptidase M32 [Paenibacillus pasadenensis]
Length=497
Score = 33.9 bits (76), Expect = 6.4, Method: Composition-based stats.
Identities = 21/59 (36%), Positives = 32/59 (54%), Gaps = 4/59 (7%)
Query 3 ALRKLRQDNDINAFRYSMERVFGNSSDVKDVASELVKRMGLLL--MRPD--IKELDQMF 57
A + R+DND AF+ +ERV G ++ D+ R LL PD ++ELD++F
Sbjct 112 AWEQARKDNDFPAFQGYLERVIGYTNQFIDLWGPKATRYDTLLDDYEPDMTVQELDEIF 170
Lambda K H a alpha
0.321 0.136 0.371 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 430540141590