bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: All non-redundant GenBank CDS translations+PDB+SwissProt+PIR+PRF
excluding environmental samples from WGS projects
49,011,213 sequences; 17,563,301,199 total letters
Query= Contig-32_CDS_annotation_glimmer3.pl_2_3
Length=544
Score E
Sequences producing significant alignments: (Bits) Value
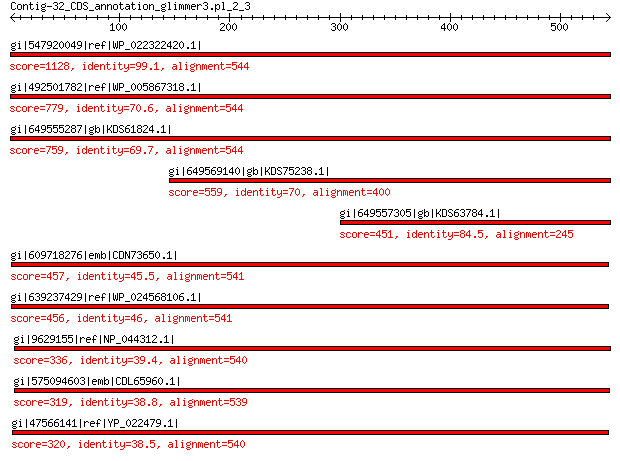
gi|547920049|ref|WP_022322420.1| capsid protein VP1 1128 0.0
gi|492501782|ref|WP_005867318.1| hypothetical protein 779 0.0
gi|649555287|gb|KDS61824.1| capsid family protein 759 0.0
gi|649569140|gb|KDS75238.1| capsid family protein 559 0.0
gi|649557305|gb|KDS63784.1| capsid family protein 451 1e-153
gi|609718276|emb|CDN73650.1| conserved hypothetical protein 457 2e-151
gi|639237429|ref|WP_024568106.1| hypothetical protein 456 4e-151
gi|9629155|ref|NP_044312.1| VP1 336 6e-104
gi|575094603|emb|CDL65960.1| unnamed protein product 319 2e-98
gi|47566141|ref|YP_022479.1| structural protein 320 3e-98
>gi|547920049|ref|WP_022322420.1| capsid protein VP1 [Parabacteroides merdae CAG:48]
gi|524592961|emb|CDD13573.1| capsid protein VP1 [Parabacteroides merdae CAG:48]
Length=553
Score = 1128 bits (2918), Expect = 0.0, Method: Compositional matrix adjust.
Identities = 539/544 (99%), Positives = 540/544 (99%), Gaps = 0/544 (0%)
Query 1 MKRPRRNAFNLSYESKLTLNMGELVPIMCMPVVPGDKFRVKTESLVRLAPLVAPMMHRVN 60
MKRPRRNAFNLSYESKLTLNMGELVPIMCMPVV GDKFRVKTESLVRLAPLVAPMMHRVN
Sbjct 10 MKRPRRNAFNLSYESKLTLNMGELVPIMCMPVVSGDKFRVKTESLVRLAPLVAPMMHRVN 69
Query 61 VFTHYFFVPNRLVWNEWEDFITKGVDGEDMPMFPKIQINQDSHLVSSASLIKEYFGDSSL 120
VFTHYFFVPNRLVWNEWEDFITKGVDGEDMPMFPKIQINQDSHLVSSASLIKEYFGDSSL
Sbjct 70 VFTHYFFVPNRLVWNEWEDFITKGVDGEDMPMFPKIQINQDSHLVSSASLIKEYFGDSSL 129
Query 121 WDYLGLPTLSACGNKSYDVVNGVKVPSGFQVSALPFRAYQLIYNEYYRDQNLTEPIDFTL 180
WDYLGLPTLSACGNKSYDVVNGVKVPSGFQVSALPFRAYQLIYNEYYRDQNLTEPIDFTL
Sbjct 130 WDYLGLPTLSACGNKSYDVVNGVKVPSGFQVSALPFRAYQLIYNEYYRDQNLTEPIDFTL 189
Query 181 GSGTTVGGDQLMALMSLRRRAWEKDYFTSALPWLQRGPEVTVPVQGAGGSMDVVYERQSD 240
GSGTTVGGDQLMALMSLRRRAWEKDYFTSALPWLQRGPEVTVPVQGAGGSMDVVYERQSD
Sbjct 190 GSGTTVGGDQLMALMSLRRRAWEKDYFTSALPWLQRGPEVTVPVQGAGGSMDVVYERQSD 249
Query 241 SQKWVDSSGREFENGRAYDITMTRANDPNSALMVAVNGGTNNRAPELDPNGTLKVNVDEM 300
SQKWVDSSGREFENG AYDITM RANDPNSALMVAVNGGTNNRAPELDPNGTLKVNVDEM
Sbjct 250 SQKWVDSSGREFENGHAYDITMARANDPNSALMVAVNGGTNNRAPELDPNGTLKVNVDEM 309
Query 301 GININDLRTSNALQRWFERNARGGSRYIEQILSHFGVRSSDARLQRPQFLGGGRMPISVS 360
GININDLRTSNALQRWFERNARGGSRYIEQILSHFGVRSSDARLQRPQFLGGGRMPISVS
Sbjct 310 GININDLRTSNALQRWFERNARGGSRYIEQILSHFGVRSSDARLQRPQFLGGGRMPISVS 369
Query 361 EVLQTSSTDETSPQANMAGHGISAGINNGFKHYFEEHGYIIGIMSITPRSGYQQGVPRDF 420
EVLQTSSTDETSPQANMAGHGISAGINNGFKHYFEEHGYIIGIMSITPRSGYQQGVPRDF
Sbjct 370 EVLQTSSTDETSPQANMAGHGISAGINNGFKHYFEEHGYIIGIMSITPRSGYQQGVPRDF 429
Query 421 TKFDNMDFYFPEFAHLSEQEIKNQELFVSEDAAYNNGTFGYTPRYAEYKYHPSEAHGDFR 480
TKFDNMDFYFPEFAHLSEQEIKNQELFVSEDAAYNNGTFGYTPRYAEYKYHPSEAHGDFR
Sbjct 430 TKFDNMDFYFPEFAHLSEQEIKNQELFVSEDAAYNNGTFGYTPRYAEYKYHPSEAHGDFR 489
Query 481 SNLSFWHLNRIFEDKPNLNTTFVECRPSNRVFATSETEDDKFWVQMYQDVKALRLMPKYG 540
NLSFWHLNRIFEDKPNLNTTFVEC+PSNRVFATSETEDDKFWVQMYQDVKALRLMPKYG
Sbjct 490 GNLSFWHLNRIFEDKPNLNTTFVECKPSNRVFATSETEDDKFWVQMYQDVKALRLMPKYG 549
Query 541 TPML 544
TPML
Sbjct 550 TPML 553
>gi|492501782|ref|WP_005867318.1| hypothetical protein [Parabacteroides distasonis]
gi|409230408|gb|EKN23272.1| hypothetical protein HMPREF1059_03257 [Parabacteroides distasonis
CL09T03C24]
Length=538
Score = 779 bits (2011), Expect = 0.0, Method: Compositional matrix adjust.
Identities = 384/550 (70%), Positives = 444/550 (81%), Gaps = 27/550 (5%)
Query 1 MKRPRRNAFNLSYESKLTLNMGELVPIMCMPVVPGDKFRVKTESLVRLAPLVAPMMHRVN 60
+KRPRRN FNLSYE+KLT N GELVPIMC PVVPGDKFRV TE LVRLAPLVAPMMHRV+
Sbjct 10 LKRPRRNVFNLSYENKLTANAGELVPIMCKPVVPGDKFRVNTEMLVRLAPLVAPMMHRVD 69
Query 61 VFTHYFFVPNRLVWNEWEDFITKGVDGEDMPMFPKIQINQDSHLVSSASLIKEYFGDSSL 120
VFTHYFFVPNRL+WN+WEDFITKGVDG D P+FPKI + D +SA+++ + D SL
Sbjct 70 VFTHYFFVPNRLLWNQWEDFITKGVDGTDTPVFPKIALRPDWVNPTSAAVLLD---DGSL 126
Query 121 WDYLGLPTLSACGNKSYD--VVNGVKVPSGFQVSALPFRAYQLIYNEYYRDQNLTEPIDF 178
WDYLGLPT+ N ++ N V P G+QVSALPFRAYQLIYNEYYRDQNLT+PI+F
Sbjct 127 WDYLGLPTIGGFNNVAFPNRSPNSVMPPVGYQVSALPFRAYQLIYNEYYRDQNLTKPIEF 186
Query 179 TLGSGTTVGGDQLMALMSLRRRAWEKDYFTSALPWLQRGPEVTVPVQGAGGSMDVVYERQ 238
+L SG + D++ L++LRRR WEKDYFTSALPW+QRGPEVTVP+QG+GG++DV +
Sbjct 187 SLNSGIVLSADEVTRLLTLRRRTWEKDYFTSALPWVQRGPEVTVPIQGSGGNLDVTLKND 246
Query 239 SDSQKWVDSSGREFENGRAYDITMTRANDPNSALMVA----VNGGTNNRAPELDPNGTLK 294
+ + Y + T +N P A+ + + GGT+ E D +
Sbjct 247 AHAD--------------TYRMPGT-SNRPAGAMQLVGGALIAGGTDGAYLEPD---NFQ 288
Query 295 VNVDEMGININDLRTSNALQRWFERNARGGSRYIEQILSHFGVRSSDARLQRPQFLGGGR 354
VNVDE+G++INDLRTSNALQRWFERNAR GSRYIEQILSHFGVRSSDARLQRPQFLGGGR
Sbjct 289 VNVDELGVSINDLRTSNALQRWFERNARSGSRYIEQILSHFGVRSSDARLQRPQFLGGGR 348
Query 355 MPISVSEVLQTSSTDETSPQANMAGHGISAGINNGFKHYFEEHGYIIGIMSITPRSGYQQ 414
PISVSEVLQTS+TD TSPQANMAGHGISAG+N+GFK YFEEHGYIIGIMSI PR+GYQQ
Sbjct 349 TPISVSEVLQTSATDSTSPQANMAGHGISAGVNHGFKRYFEEHGYIIGIMSIRPRTGYQQ 408
Query 415 GVPRDFTKFDNMDFYFPEFAHLSEQEIKNQELFVSEDAAYNNGTFGYTPRYAEYKYHPSE 474
GVP+DF KFDNMDFYFPEFAHL EQEIKN+E+++ + A NNGTFGYTPRYAEYKY +E
Sbjct 409 GVPKDFRKFDNMDFYFPEFAHLGEQEIKNEEVYLQQTPASNNGTFGYTPRYAEYKYSMNE 468
Query 475 AHGDFRSNLSFWHLNRIFEDKPNLNTTFVECRPSNRVFATSETEDDKFWVQMYQDVKALR 534
HGDFR N++FWHLNRIF + PNLNTTFVEC PSNRVFAT+ET DDK+W+Q+YQDVKALR
Sbjct 469 VHGDFRGNMAFWHLNRIFSESPNLNTTFVECNPSNRVFATAETSDDKYWIQLYQDVKALR 528
Query 535 LMPKYGTPML 544
LMPKYGTPML
Sbjct 529 LMPKYGTPML 538
>gi|649555287|gb|KDS61824.1| capsid family protein [Parabacteroides distasonis str. 3999B
T(B) 4]
gi|649560568|gb|KDS66876.1| capsid family protein [Parabacteroides distasonis str. 3999B
T(B) 4]
gi|649561020|gb|KDS67307.1| capsid family protein [Parabacteroides distasonis str. 3999B
T(B) 4]
gi|649562724|gb|KDS68908.1| capsid family protein [Parabacteroides distasonis str. 3999B
T(B) 6]
Length=541
Score = 759 bits (1961), Expect = 0.0, Method: Compositional matrix adjust.
Identities = 379/549 (69%), Positives = 432/549 (79%), Gaps = 22/549 (4%)
Query 1 MKRPRRNAFNLSYESKLTLNMGELVPIMCMPVVPGDKFRVKTESLVRLAPLVAPMMHRVN 60
+KRPRRN FNLSYE+KLT+N GEL+PIMC PVVPGDKFRV TE LVRLAPLVAPMMHRV+
Sbjct 10 LKRPRRNVFNLSYENKLTVNAGELIPIMCKPVVPGDKFRVNTEMLVRLAPLVAPMMHRVD 69
Query 61 VFTHYFFVPNRLVWNEWEDFITKGVDGEDMPMFPKIQINQDSHLVSSASLIKEYFGDSSL 120
VFTHYFFVPNRL+WN+WEDFITKGVDG D P+FP V +A+ FGD SL
Sbjct 70 VFTHYFFVPNRLIWNKWEDFITKGVDGTDSPVFPTYSF---PSTVDTAN-AHNSFGDGSL 125
Query 121 WDYLGLPTLSACGNKSYDVV--NGVKVPSGFQVSALPFRAYQLIYNEYYRDQNLTEPIDF 178
WDYLGLP+++ G + V NGVK P+GF+VSALPFRAY LIYNEYYRDQNLT ++
Sbjct 126 WDYLGLPSINQIGEAVFQVQSPNGVKAPAGFKVSALPFRAYHLIYNEYYRDQNLTSELEI 185
Query 179 TLGSGTTVGGDQL---MALMSLRRRAWEKDYFTSALPWLQRGPEVTVPVQGAGGSMDVVY 235
TL SG QL +L L RRAWEKDYFTSALPW+QRGPEVTVP+ G GG + V
Sbjct 186 TLDSGNY----QLPVNSSLWQLHRRAWEKDYFTSALPWVQRGPEVTVPING-GGEIPVEM 240
Query 236 ERQSDSQKWVDSSGREFENGRAYDITMTRANDPNSALMVAVNGGTNNRAPELDPNGTLKV 295
+ +QK R+ +G + S L G +A ++P+ + V
Sbjct 241 KEGFAAQKITTFPDRKPISGSEVLYSAP------SVLSYGQIGSIKGQA-LIEPDNFV-V 292
Query 296 NVDEMGININDLRTSNALQRWFERNARGGSRYIEQILSHFGVRSSDARLQRPQFLGGGRM 355
N D+MG+NIND+RTSNALQRWFERNAR GSRYIEQILSHFGVRSSDARLQRPQFLGGGR
Sbjct 293 NTDQMGVNINDIRTSNALQRWFERNARSGSRYIEQILSHFGVRSSDARLQRPQFLGGGRT 352
Query 356 PISVSEVLQTSSTDETSPQANMAGHGISAGINNGFKHYFEEHGYIIGIMSITPRSGYQQG 415
PISVSEVLQTSSTD TSPQANMAGHGISAG+N+GF YFEEHGYI+GIMSI PR+GYQQG
Sbjct 353 PISVSEVLQTSSTDSTSPQANMAGHGISAGVNHGFTRYFEEHGYIMGIMSIRPRTGYQQG 412
Query 416 VPRDFTKFDNMDFYFPEFAHLSEQEIKNQELFVSEDAAYNNGTFGYTPRYAEYKYHPSEA 475
VP+DF KFDNMDFYFPEFAHL EQEIKN+EL+++E A N GTFGYTPRYAEYKY +E
Sbjct 413 VPKDFRKFDNMDFYFPEFAHLGEQEIKNEELYLNESDAANEGTFGYTPRYAEYKYSQNEV 472
Query 476 HGDFRSNLSFWHLNRIFEDKPNLNTTFVECRPSNRVFATSETEDDKFWVQMYQDVKALRL 535
HGDFR N++FWHLNRIF++KPNLNTTFVEC PSNRVFAT+ET DDK+WVQ+YQD+KALRL
Sbjct 473 HGDFRGNMAFWHLNRIFKEKPNLNTTFVECNPSNRVFATAETSDDKYWVQIYQDIKALRL 532
Query 536 MPKYGTPML 544
MPKYGTPML
Sbjct 533 MPKYGTPML 541
>gi|649569140|gb|KDS75238.1| capsid family protein, partial [Parabacteroides distasonis str.
3999B T(B) 6]
Length=390
Score = 559 bits (1441), Expect = 0.0, Method: Compositional matrix adjust.
Identities = 280/403 (69%), Positives = 319/403 (79%), Gaps = 16/403 (4%)
Query 145 VPSGFQVSALPFRAYQLIYNEYYRDQNLTEPIDFTLGSGTTVGGDQL---MALMSLRRRA 201
P+GF+VSALPFRAY LIYNEYYRDQNLT ++ TL SG QL +L L RRA
Sbjct 1 APAGFKVSALPFRAYHLIYNEYYRDQNLTSELEITLDSGNY----QLPVNSSLWQLHRRA 56
Query 202 WEKDYFTSALPWLQRGPEVTVPVQGAGGSMDVVYERQSDSQKWVDSSGREFENGRAYDIT 261
WEKDYFTSALPW+QRGPEVTVP+ G GG + V + +QK R+ +G +
Sbjct 57 WEKDYFTSALPWVQRGPEVTVPING-GGEIPVEMKEGFAAQKITTFPDRKPISGSEVLYS 115
Query 262 MTRANDPNSALMVAVNGGTNNRAPELDPNGTLKVNVDEMGININDLRTSNALQRWFERNA 321
S L G +A ++P+ + VN D+MG+NIND+RTSNALQRWFERNA
Sbjct 116 AP------SVLSYGQIGSIKGQA-LIEPDNFV-VNTDQMGVNINDIRTSNALQRWFERNA 167
Query 322 RGGSRYIEQILSHFGVRSSDARLQRPQFLGGGRMPISVSEVLQTSSTDETSPQANMAGHG 381
R GSRYIEQILSHFGVRSSDARLQRPQFLGGGR PISVSEVLQTSSTD TSPQANMAGHG
Sbjct 168 RSGSRYIEQILSHFGVRSSDARLQRPQFLGGGRTPISVSEVLQTSSTDSTSPQANMAGHG 227
Query 382 ISAGINNGFKHYFEEHGYIIGIMSITPRSGYQQGVPRDFTKFDNMDFYFPEFAHLSEQEI 441
ISAG+N+GF YFEEHGYI+GIMSI PR+GYQQGVP+DF KFDNMDFYFPEFAHL EQEI
Sbjct 228 ISAGVNHGFTRYFEEHGYIMGIMSIRPRTGYQQGVPKDFRKFDNMDFYFPEFAHLGEQEI 287
Query 442 KNQELFVSEDAAYNNGTFGYTPRYAEYKYHPSEAHGDFRSNLSFWHLNRIFEDKPNLNTT 501
KN+EL+++E A N GTFGYTPRYAEYKY +E HGDFR N++FWHLNRIF++KPNLNTT
Sbjct 288 KNEELYLNESDAANEGTFGYTPRYAEYKYSQNEVHGDFRGNMAFWHLNRIFKEKPNLNTT 347
Query 502 FVECRPSNRVFATSETEDDKFWVQMYQDVKALRLMPKYGTPML 544
FVEC PSNRVFAT+ET DDK+WVQ+YQD+KALRLMPKYGTPML
Sbjct 348 FVECNPSNRVFATAETSDDKYWVQIYQDIKALRLMPKYGTPML 390
>gi|649557305|gb|KDS63784.1| capsid family protein [Parabacteroides distasonis str. 3999B
T(B) 4]
gi|649559156|gb|KDS65543.1| capsid family protein [Parabacteroides distasonis str. 3999B
T(B) 6]
Length=245
Score = 451 bits (1161), Expect = 1e-153, Method: Compositional matrix adjust.
Identities = 207/245 (84%), Positives = 227/245 (93%), Gaps = 0/245 (0%)
Query 300 MGININDLRTSNALQRWFERNARGGSRYIEQILSHFGVRSSDARLQRPQFLGGGRMPISV 359
MG+NIND+RTSNALQRWFERNAR GSRYIEQILSHFGVRSSDARLQRPQFLGGGR PISV
Sbjct 1 MGVNINDIRTSNALQRWFERNARSGSRYIEQILSHFGVRSSDARLQRPQFLGGGRTPISV 60
Query 360 SEVLQTSSTDETSPQANMAGHGISAGINNGFKHYFEEHGYIIGIMSITPRSGYQQGVPRD 419
SEVLQTSSTD TSPQANMAGHGISAG+N+GF YFEEHGYI+GIMSI PR+GYQQGVP+D
Sbjct 61 SEVLQTSSTDSTSPQANMAGHGISAGVNHGFTRYFEEHGYIMGIMSIRPRTGYQQGVPKD 120
Query 420 FTKFDNMDFYFPEFAHLSEQEIKNQELFVSEDAAYNNGTFGYTPRYAEYKYHPSEAHGDF 479
F KFDNMDFYFPEFAHL EQEIKN+EL+++E A N GTFGYTPRYAEYKY +E HGDF
Sbjct 121 FRKFDNMDFYFPEFAHLGEQEIKNEELYLNESDAANEGTFGYTPRYAEYKYSQNEVHGDF 180
Query 480 RSNLSFWHLNRIFEDKPNLNTTFVECRPSNRVFATSETEDDKFWVQMYQDVKALRLMPKY 539
R N++FWHLNRIF++KPNLNTTFVEC PSNRVFAT+ET DDK+WVQ+YQD+KALRLMPKY
Sbjct 181 RGNMAFWHLNRIFKEKPNLNTTFVECNPSNRVFATAETSDDKYWVQIYQDIKALRLMPKY 240
Query 540 GTPML 544
GTPML
Sbjct 241 GTPML 245
>gi|609718276|emb|CDN73650.1| conserved hypothetical protein [Elizabethkingia anophelis]
Length=537
Score = 457 bits (1175), Expect = 2e-151, Method: Compositional matrix adjust.
Identities = 246/568 (43%), Positives = 341/568 (60%), Gaps = 71/568 (13%)
Query 2 KRPRRNAFNLSYESKLTLNMGELVPIMCMPVVPGDKFRVKTESLVRLAPLVAPMMHRVNV 61
K P+ + FN+SY+ K ++N G+LVPI C V+PGDK + + + RLAP++AP+MH VNV
Sbjct 10 KAPKSSTFNMSYDRKFSMNFGDLVPIHCQEVIPGDKISINPQHMTRLAPMIAPVMHEVNV 69
Query 62 FTHYFFVPNRLVWNEWEDFITKGVDGEDMPMFPKIQINQDSHLVSSASLIKEYFGDSSLW 121
F HYFFVPNR++W+ WE FIT G G D + P++ +L S SL
Sbjct 70 FIHYFFVPNRIIWSNWEQFITGGESGLDQHLMPRV-----GNLPVSKG---------SLA 115
Query 122 DYLGLPTLS---ACGNKS--YDVVNGVKVPSGFQVSALPFRAYQLIYNEYYRDQNLTEPI 176
D+LGLP + A GN Y++VN LPF AYQ I++E+YRD+NL +P+
Sbjct 116 DHLGLPLTTGRFAVGNAGVLYNLVN-----------LLPFLAYQKIWDEFYRDENLIQPL 164
Query 177 DFTLGSGTTVGG-------------DQLMALMSLRRRAWEKDYFTSALPWLQRGPEVTVP 223
F +G V + L +R+RAW DYFTSALP+ Q+G V +P
Sbjct 165 -FRDSNGNPVKMFNDGINDHNLPPYSKFTELFKMRKRAWHHDYFTSALPFAQKGNAVKIP 223
Query 224 VQGAGGSMDVVYERQSDSQKWVDSSGREFENGRAYDITMTRANDPNSALMVAVNGGTNNR 283
+ G++ + YE S + + M PN L VNG +
Sbjct 224 I-FPQGNVPLTYEMGS----------------QTFIKDMAGNPAPNKDLRSDVNGNLQDV 266
Query 284 APE---LDPNGTLKVNVDEMGIN-INDLRTSNALQRWFERNARGGSRYIEQILSHFGVRS 339
+ + LDP+ LK+N+ ++ +NDLR + LQ W E+NAR GSRY E ILS FGV++
Sbjct 267 SGQPLSLDPSKNLKLNMASENVSTVNDLRRAFKLQEWLEKNARAGSRYAESILSFFGVKT 326
Query 340 SDARLQRPQFLGGGRMPISVSEVLQTSSTDETSPQANMAGHGISAGINNGFKHYFEEHGY 399
SD RLQRP+FLGG + PI +SEVLQ S+TD T+PQ NMAGHGI G + GF +FEEHGY
Sbjct 327 SDGRLQRPEFLGGNKSPIMISEVLQQSATDSTTPQGNMAGHGIGIGKDGGFSRFFEEHGY 386
Query 400 IIGIMSITPRSGYQQGVPRDFTKFDNMDFYFPEFAHLSEQEIKNQELFVSE-DAAYNNGT 458
+IG+MS+ P++ Y QG+PR F+K D D+++P+F H+ EQ + N+E+F DA +
Sbjct 387 VIGLMSVIPKTSYSQGIPRHFSKSDKFDYFWPQFEHIGEQPVYNKEIFAKNIDAFDSEAV 446
Query 459 FGYTPRYAEYKYHPSEAHGDFRSNLSFWHLNRIFE-DKPN-LNTTFVECRPS--NRVFAT 514
FGY PRY+EYK+ PS HGDF+ +L FWHL RIF+ DKP LN +F+EC + +R+FA
Sbjct 447 FGYLPRYSEYKFSPSTVHGDFKDDLYFWHLGRIFDTDKPPVLNQSFIECDKNALSRIFAV 506
Query 515 SETEDDKFWVQMYQDVKALRLMPKYGTP 542
E + DKF+ +YQ + A R M +G P
Sbjct 507 -EDDTDKFYCHLYQKITAKRKMSYFGDP 533
>gi|639237429|ref|WP_024568106.1| hypothetical protein [Elizabethkingia anophelis]
Length=546
Score = 456 bits (1174), Expect = 4e-151, Method: Compositional matrix adjust.
Identities = 249/567 (44%), Positives = 335/567 (59%), Gaps = 60/567 (11%)
Query 2 KRPRRNAFNLSYESKLTLNMGELVPIMCMPVVPGDKFRVKTESLVRLAPLVAPMMHRVNV 61
K P+ + FN+SY+ K ++N G+LVPI C +VPGDK + + + RLAP++AP+MH VNV
Sbjct 10 KAPKSSTFNMSYDRKFSMNFGDLVPIHCQEIVPGDKISINPQHMTRLAPMLAPVMHEVNV 69
Query 62 FTHYFFVPNRLVWNEWEDFITKGVDGEDMPMFPKIQINQDSHLVSSASLIKEYFGDSSLW 121
F HYFFVPNR++W WE FIT G G D M P +Q + + K SSL
Sbjct 70 FIHYFFVPNRILWKNWEAFITGGQSGLDAHMLPVVQ---------NLPVPK-----SSLG 115
Query 122 DYLGLPTLSACGNKSYDVVNGVKVPSGFQVSALPFRAYQLIYNEYYRDQNLTEPIDFTLG 181
DYLGLP + V N +P +VS LPF AYQ I++EYYRD+NL + + F
Sbjct 116 DYLGLPLTEG----RFAVGNDGVLPD--RVSMLPFLAYQKIWDEYYRDENLIDSV-FVDK 168
Query 182 SGTT----VGGD---------QLMALMSLRRRAWEKDYFTSALPWLQRGPEVTVPVQ--- 225
+G + G + L +++RAW DYFTSALP+ Q+G V +P+Q
Sbjct 169 NGDKRELFIDGINYWNPSLPYEFRQLFDIKKRAWHHDYFTSALPFAQKGAAVKMPLQMTA 228
Query 226 ----GAGGSMDVVYERQSDSQKWVDSSGREFENGRAYDITMTRANDPNSALMVAVNGGTN 281
GG+ + ++ D + +G E+G D LMV + N
Sbjct 229 DLFYNPGGN---TFVKKPDGS--LSHTGFRLEDGSV-------PADGIGHLMVETSSTGN 276
Query 282 NRAPELDPNGTLKVNVDEM-GININDLRTSNALQRWFERNARGGSRYIEQILSHFGVRSS 340
+ +D + L V++ G INDLR + LQ W E+NAR GSRY E ILS FGV++S
Sbjct 277 SNPVNIDNSSNLGVDLKTASGSTINDLRRAFKLQEWLEKNARAGSRYAESILSFFGVKTS 336
Query 341 DARLQRPQFLGGGRMPISVSEVLQTSSTDETSPQANMAGHGISAGINNGFKHYFEEHGYI 400
D RLQRP+FLGG + PI +SEVLQ SSTD T+PQ NMAGHGIS G GF +FEEHGY+
Sbjct 337 DGRLQRPEFLGGNKTPILISEVLQQSSTDSTTPQGNMAGHGISVGKEGGFSKFFEEHGYV 396
Query 401 IGIMSITPRSGYQQGVPRDFTKFDNMDFYFPEFAHLSEQEIKNQELFVSEDAAYNN-GTF 459
IG+MS+ P++ Y QG+PR F+KFD D+++P+F H+ EQ + N+E+F Y++ G F
Sbjct 397 IGLMSVIPKTSYSQGIPRHFSKFDKFDYFWPQFEHIGEQPVYNKEIFAKNVGDYDSGGVF 456
Query 460 GYTPRYAEYKYHPSEAHGDFRSNLSFWHLNRIFEDK--PNLNTTFVECRPS--NRVFATS 515
GY PRY+EYKY PS HGDF+ L FWHL RIF+ P LN F+E S +R+FA
Sbjct 457 GYVPRYSEYKYSPSTIHGDFKDTLYFWHLGRIFDSSAPPKLNRDFIEVNKSGLSRIFAV- 515
Query 516 ETEDDKFWVQMYQDVKALRLMPKYGTP 542
E DKF+ +YQ + A R M +G P
Sbjct 516 EDNSDKFYCHLYQKITAKRKMSYFGDP 542
>gi|9629155|ref|NP_044312.1| VP1 [Chlamydia phage 1]
gi|139180|sp|P19192.2|F_BPCHP RecName: Full=Capsid protein VP1; AltName: Full=Protein VP1;
Short=VP1 [Chlamydia phage 1]
gi|93817|pir||JU0345 major capsid protein VP1 - Chlamydophila psittaci phage Chp1
gi|217762|dbj|BAA00515.1| VP1 [Chlamydia phage 1]
Length=596
Score = 336 bits (862), Expect = 6e-104, Method: Compositional matrix adjust.
Identities = 213/593 (36%), Positives = 300/593 (51%), Gaps = 76/593 (13%)
Query 5 RRNAFNLSYESKLTLNMGELVPIMCMPVVPGDKFRVKTESLVRLAPLVAPMMHRVNVFTH 64
RR++F+ S+ K T +M LVP V+PGD F + L RL LV P+M + + T
Sbjct 24 RRSSFDRSHGYKTTFDMDYLVPFFVDEVLPGDTFSLSETHLCRLTTLVQPIMDNIQLTTQ 83
Query 65 YFFVPNRLVWNEWEDFITKGVDGEDMPMFPKIQINQDSHLVSSASLIKEYFGDSSLWDYL 124
+FFVPNRL+W+ WE FIT G D P+ + + V + + ++S++DY
Sbjct 84 FFFVPNRLLWDNWESFITGG----DEPVAWTSTNPANEYFVPQVTSPDGGYAENSIYDYF 139
Query 125 GLPTLSACGNKSYDVVNGVKVPSGFQVSALPFRAYQLIYNEYYRDQNLTEPIDFTLGSGT 184
GLPT A ++ LP RAY LI+NEYYRD+NL E + G
Sbjct 140 GLPTKVA----------------NYRHQVLPLRAYNLIFNEYYRDENLQESLPVWTGDAD 183
Query 185 -----TVG-----GDQLMALMSLRRRAWEKDYFTSALPWLQRGPEVTVPVQGA------- 227
T G D + + L RR DYFTSALP LQ+GP V + + G
Sbjct 184 PKVDPTTGEESQEDDAVPYVYKLMRRNKRYDYFTSALPGLQKGPSVGIGITGGDSGRLPV 243
Query 228 -GGSMDVVYERQSD------------SQKWVDSSGREFENGRAYDITMTRANDPNSALM- 273
G ++ + SD SQKW + GR +G T N P ++
Sbjct 244 HGLAIRSYLDDSSDDQFSFGVSYVNASQKWFTADGR-LTSGMGSVPVGTTGNFPIDNVVY 302
Query 274 -------VAVNGGTNNRAPELDPNGTLKVNVD---EMGININDLRTSNALQRWFERNARG 323
VA G ++ + G V VD + IN LR + LQ+WFE++AR
Sbjct 303 PSYFGTTVAQTGSPSSSSTPPFVKGDFPVYVDLAASSSVTINSLRNAITLQQWFEKSARY 362
Query 324 GSRYIEQILSHFGVRSSDARLQRPQFLGGGRMPISVSEVLQTSSTDETSPQANMAGHGIS 383
GSRY+E + HFGV D R QRP +LGG + +SV+ V+Q SSTD SPQ N++ + +S
Sbjct 363 GSRYVESVQGHFGVHLGDYRAQRPIYLGGSKSYVSVNPVVQNSSTDSVSPQGNLSAYALS 422
Query 384 AGINNGFKHYFEEHGYIIGIMSITPRSGYQQGVPRDFTKFDNMDFYFPEFAHLSEQEIKN 443
+ F F EHG++IG++S T YQQG+ R +++F D+Y+P FAHL EQ + N
Sbjct 423 TDTKHLFTKSFVEHGFVIGLLSATADLTYQQGLERQWSRFSRYDYYWPTFAHLGEQPVYN 482
Query 444 QELFVSED-------AAYNNGTFGYTPRYAEYKYHPSEAHGDFRSN----LSFWHLNRIF 492
+E++ D +A N+ FGY RYAEY+Y PS+ G FRSN L WHL++ F
Sbjct 483 KEIYCQSDTVMDPSGSAVNDVPFGYQERYAEYRYKPSKVTGLFRSNATGTLDSWHLSQNF 542
Query 493 EDKPNLNTTFVECR-PSNRVFATSETEDDKFWVQMYQDVKALRLMPKYGTPML 544
+ P LN TF++ P +R A + D F Y + + +R MP Y P L
Sbjct 543 ANLPTLNETFIQSNTPIDRALAVPDQPD--FICDFYFNYRCIRPMPVYSVPGL 593
>gi|575094603|emb|CDL65960.1| unnamed protein product [uncultured bacterium]
Length=507
Score = 319 bits (817), Expect = 2e-98, Method: Compositional matrix adjust.
Identities = 209/558 (37%), Positives = 298/558 (53%), Gaps = 72/558 (13%)
Query 5 RRNAFNLSYESKLTLNMGELVPIMCMPVVPGDKFRVKTESLVRLAPLVAPMMHRVNVFTH 64
+R+ +LS+ + LT MG + PI C+ V+PGD F +R PLVAP+MH V+V
Sbjct 3 KRSIHSLSHFNLLTTQMGLITPIECVDVLPGDSFMQDNSVFLRTMPLVAPVMHPVHVEIR 62
Query 65 YFFVPNRLVWNEWEDFITKGVDGEDMPMFPKIQINQDSHLVSSASLIKEYFGDSSLWDYL 124
FFVP R++W+E+EDFIT G G + + P H V+ K G+ L DYL
Sbjct 63 SFFVPLRIIWDEFEDFITGGPKGLNNSVHP--------HFVADEQNCK--LGE--LNDYL 110
Query 125 GLPTLSACGNKSYDVVNGVKVPSGFQVSALPFRAYQLIYNEYYRDQNLTE--PIDFTLGS 182
G+P + G K V+ L RAY LIYNEY+RDQ++ E PI F G
Sbjct 111 GIPPDTLLGTK---------------VNCLYARAYALIYNEYFRDQDIDEELPISFESGQ 155
Query 183 GTTVGGDQLMALMSLRRRAWEKDYFTSALPWLQRGPEVTVPVQGAGGSMDVVYERQ---- 238
TT L+R W KDYFT A P+ Q+GP VP+ +GG++ +
Sbjct 156 DTTTPS-------VLQRADWRKDYFTLARPFEQKGPAAVVPITSSGGAITGLTGTTTTTV 208
Query 239 ---SDSQKWVDS----SGREFENGRAYDIT----MTRANDPNSALMVAVNGGTNNRAPEL 287
SDS +W + SG N R+ T++N ++A V+ + N + +L
Sbjct 209 TIGSDSGRWNPATGLKSGAGVNNSRSLIFGDGDGFTQSNHTHAATAVSNTVVSGNASFDL 268
Query 288 DPNGTLKVNVDEMGININDLRTSNALQRWFERNARGGSRYIEQILSHFGVRSSDARLQRP 347
GT+ +I+D R + LQR+ E + GSRY E + GVRSSD RLQ P
Sbjct 269 ---GTM---------SISDFREAMQLQRFEEIRSLYGSRY-EDYQAVLGVRSSDGRLQNP 315
Query 348 QFLGGGRMPISVSEVLQTSSTDETSPQANMAGHGISAGINNGFKHYFEEHGYIIGIMSIT 407
++LGGG+ I SEVLQTS + +SP + GHGI A F +F EHG ++ ++ +
Sbjct 316 EYLGGGQRTIQFSEVLQTS--EGSSPVGTLRGHGIGALKTKRFLRFFNEHGILLTLLVVR 373
Query 408 PRSGYQQGVPRDFTKFDNMDFYFPEFAHLSEQEIKNQELFVSEDAAYNNG--TFGYTPRY 465
P S Y QG+ R F + D++ P+F H+ +Q + N+EL+ AA NG TFG++ RY
Sbjct 374 PISIYTQGLNRMFLRETRFDYWQPQFEHIGQQSVLNKELY----AASPNGDDTFGFSNRY 429
Query 466 AEYKYHPSEAHGDFRSNLSFWHLNRIFEDKPNLNTTFVECRPSNRVFATSETEDDKFWVQ 525
EY+YHPS HG+FR+ WHL R FE P LN+ F++C P+ R+FA + D+ V
Sbjct 430 NEYRYHPSNVHGEFRTYYEDWHLGRKFESTPTLNSDFLKCHPTTRIFAETSGNYDQLLVM 489
Query 526 MYQDVKALRLMPKYGTPM 543
++A RL+ K G P+
Sbjct 490 CQNHIRARRLISKNGDPI 507
>gi|47566141|ref|YP_022479.1| structural protein [Chlamydia phage 3]
gi|47522476|emb|CAD79477.1| structural protein [Chlamydia phage 3]
Length=565
Score = 320 bits (821), Expect = 3e-98, Method: Compositional matrix adjust.
Identities = 208/579 (36%), Positives = 297/579 (51%), Gaps = 80/579 (14%)
Query 3 RPRRNAFNLSYESKLTLNMGELVPIMCMPVVPGDKFRVKTESLVRLAPLVAPMMHRVNVF 62
R +R++F+ S K T N G L+PI C V+PGD F +K L R+A + P+M + +
Sbjct 22 RIQRSSFDRSCGLKTTFNAGYLIPIFCDEVLPGDTFSLKEAFLARMATPIFPLMDNLRLD 81
Query 63 THYFFVPNRLVWNEWEDFITKGVDGEDMPMFPKIQINQDSHLVSSASLIKEYFGDSSLWD 122
T YFFVP RL+W+ ++ F + + +D F L + F + S+ D
Sbjct 82 TQYFFVPLRLIWSNFQKFCGEQDNPDDSTDF----------LTPVLTAPTGGFTEGSIHD 131
Query 123 YLGLPTLSACGNKSYDVVNGVKVPSGFQVSALPFRAYQLIYNEYYRDQNLTEPIDFTLGS 182
YLGLPT A G Q +A RAY LI+N+YYRD+N+ E ++ +G
Sbjct 132 YLGLPTKVA----------------GVQCAAFWHRAYNLIWNQYYRDENIQESVEVQMGD 175
Query 183 GTTVGGDQLMALMSLRRRAWEKDYFTSALPWLQRGPEVTVPVQGA----GGSMDV----- 233
TT D++ L +R DYFTS LPW Q+GP VT+ V G G M+V
Sbjct 176 TTT---DEVKN-YELLKRGKRYDYFTSCLPWPQKGPAVTIGVGGKAPIEGLYMNVNSNNP 231
Query 234 ----VYERQSDSQKWVDSSGREFENGRAYDITMTRANDPNSALMVAVNGGTNNRAPELDP 289
V + QS + D G + AY+ T V VN P+ +P
Sbjct 232 VGKFVLDSQSTPRVLQDLQGNKLSGIAAYNQT---------GKHVYVNSAWYTVTPQSEP 282
Query 290 NGTLKVN----------VDEMG----ININDLRTSNALQRWFERNARGGSRYIEQILSHF 335
TL+ ++G + IN LR + LQ+ +ER+ARGG+RYIE I SHF
Sbjct 283 GATLENGNYYTTQKPQIYADLGATSPVTINSLREAFQLQKLYERDARGGTRYIEIIRSHF 342
Query 336 GVRSSDARLQRPQFLGGGRMPISVSEVLQTSSTDETSPQANMAGHGISAGINNGFKHYFE 395
V+S DARLQR ++LGG P+++S + QTSSTD TSPQ N+A +G + G F F
Sbjct 343 NVQSPDARLQRAEYLGGSSTPVNISPIPQTSSTDSTSPQGNLAAYGTAIGSKRVFTKSFT 402
Query 396 EHGYIIGIMSITPRSGYQQGVPRDFTKFDNMDFYFPEFAHLSEQEIKNQELFVSEDAAYN 455
EHG I+G+ S+ YQQG+ R +++ DFY+P +HL EQ + N+E++ + N
Sbjct 403 EHGVILGLASVRADLNYQQGLDRMWSRRTRWDFYWPALSHLGEQAVLNKEIYCQGPSVKN 462
Query 456 NG-------TFGYTPRYAEYKYHPSEAHGDFRSN----LSFWHLNRIFEDKPNLNTTFVE 504
+G FGY R+AEY+Y S+ G FRSN L WHL + FE+ P L+ F+E
Sbjct 463 SGGEIVDEQVFGYQERFAEYRYKTSKITGKFRSNATSSLDSWHLAQEFENLPTLSPEFIE 522
Query 505 CRPS-NRVFATSETEDDKFWVQMYQDVKALRLMPKYGTP 542
P +RV A S + F + + ++ R MP Y P
Sbjct 523 ENPPMDRVLAVS--NEPHFLLDGWFSLRCARPMPVYSVP 559
Lambda K H a alpha
0.319 0.135 0.412 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 3935252973510