bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: All non-redundant GenBank CDS translations+PDB+SwissProt+PIR+PRF
excluding environmental samples from WGS projects
49,011,213 sequences; 17,563,301,199 total letters
Query= Contig-37_CDS_annotation_glimmer3.pl_2_2
Length=731
Score E
Sequences producing significant alignments: (Bits) Value
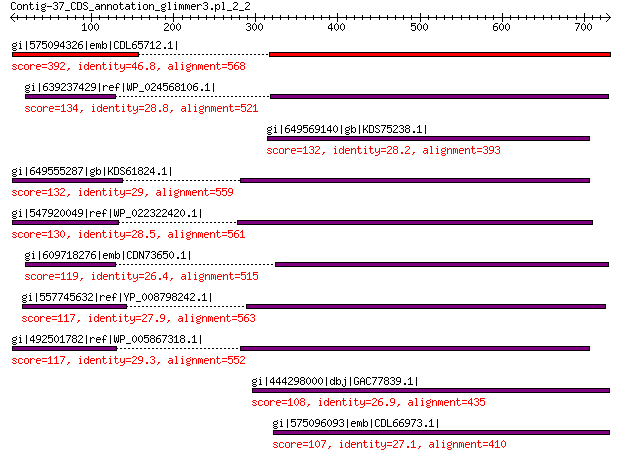
gi|575094326|emb|CDL65712.1| unnamed protein product 392 3e-121
gi|639237429|ref|WP_024568106.1| hypothetical protein 134 7e-30
gi|649569140|gb|KDS75238.1| capsid family protein 132 1e-29
gi|649555287|gb|KDS61824.1| capsid family protein 132 2e-29
gi|547920049|ref|WP_022322420.1| capsid protein VP1 130 1e-28
gi|609718276|emb|CDN73650.1| conserved hypothetical protein 119 5e-25
gi|557745632|ref|YP_008798242.1| major capsid protein 117 2e-24
gi|492501782|ref|WP_005867318.1| hypothetical protein 117 2e-24
gi|444298000|dbj|GAC77839.1| major capsid protein 108 2e-21
gi|575096093|emb|CDL66973.1| unnamed protein product 107 4e-21
>gi|575094326|emb|CDL65712.1| unnamed protein product [uncultured bacterium]
Length=758
Score = 392 bits (1008), Expect = 3e-121, Method: Compositional matrix adjust.
Identities = 194/416 (47%), Positives = 281/416 (68%), Gaps = 16/416 (4%)
Query 317 LRLLAYRFRAYESVYNAYYRDIRNNPFIVDGRPVYNKWLPSMKGGADN-TTYSLHQCNWE 375
++L AY FRAYE++YNAY R+ RNNPF+++G+ YN+W+ + GG+D T L NW+
Sbjct 358 IKLSAYPFRAYEAIYNAYIRNTRNNPFVLNGKKTYNRWITTDAGGSDTLTPRDLRFANWQ 417
Query 376 RDFLTTAVPNPQQGANAPLVGLTVGDVVTRADDGTYSIQKQTVLVDEDGSKYGISYKVSE 435
D TTA+ PQQG APLVGLT ++ + D G T +VDE+G+ Y + ++
Sbjct 418 SDAYTTALTAPQQGV-APLVGLTTYEIRSVNDAGHEVTTVNTAIVDEEGNAYKVDFE--S 474
Query 436 DGESLVGVDYDPVSEKTPVTAINSYAELAALTTTEGSGFTIETLRYVNAYQKFLELNMRK 495
+GE+L GV+Y P+ V + +L + SG +I R VNAYQ++LELN +
Sbjct 475 NGEALKGVNYTPLKAGEAV-------NMQSLVSPVTSGISINDFRNVNAYQRYLELNQFR 527
Query 496 GFSYKQIMQGRWDIDIRFDELLMPEFIGGISRELSMRTVEQTVDQQSTSSQGQYAEALGS 555
GFSYK+I++GR+D+++R+D L MPE++GGI+R++ + + QTV+ T+ G Y +LGS
Sbjct 528 GFSYKEIIEGRFDVNVRYDALNMPEYLGGITRDIVVNPITQTVE---TTGSGSYVGSLGS 584
Query 556 KSGIAGVYGSTSNNIEVFCDEESYIIGLLTVTPVPIYTQMLSKDFLYNGLLDHYQPEFDR 615
+SG+A +G+T +I VFCDEES ++G++ V P+P+Y +L K Y LD + PEFD
Sbjct 585 QSGLATCFGNTDGSISVFCDEESIVMGIMYVMPMPVYDSLLPKWLTYRERLDSFNPEFDH 644
Query 616 IGFQPITYKEVCPLNLGVADTVNKANQTFGYQRPWYEYVAKYDSAHGLFRTNMKNFVMSR 675
IG+QPI KE+ P+ V D ++ N FGYQRPWYEYVAK D AHGLF ++++NF+M R
Sbjct 645 IGYQPIYAKELGPMQC-VQDDID-PNTVFGYQRPWYEYVAKPDRAHGLFLSSLRNFIMFR 702
Query 676 VFSGLPQLGQQFLLVDPDTVNQVFSVTEYTDKIFGYVKFNATARLPISRVAIPRLD 731
F +P+LGQ F ++ P +VN VFSVTE +DKI G + F+ TA+LPISRV +PRL+
Sbjct 703 SFDNVPELGQSFTVMQPGSVNNVFSVTEVSDKILGQIHFDCTAQLPISRVVVPRLE 758
Score = 141 bits (356), Expect = 1e-31, Method: Compositional matrix adjust.
Identities = 72/158 (46%), Positives = 98/158 (62%), Gaps = 7/158 (4%)
Query 4 NIFDATFDANNRIDVNSFDWSHVNNLTTDFGRITPVFCELVPAKGSLRINPEFGLELMPM 63
++F+ D N + NSFDWSH NN TTD GRITPVF ELVP S+RI PEFGL MPM
Sbjct 3 SVFNKIGDIKNDVKRNSFDWSHDNNFTTDLGRITPVFTELVPPNSSIRIKPEFGLRFMPM 62
Query 64 VFPVQTRMFARLNFFKVTLRSMWEDYSDFISNFRDDLEE---PYILPGSQNFTT--MLGT 118
+FP+QT+M A L+F+KV LR++W DY DFIS+ D+ EE PY+ S +++ L
Sbjct 63 MFPIQTKMKAYLSFYKVPLRTLWADYMDFISS--DNTEEFQPPYMSFDSTDYSEGGTLAP 120
Query 119 GSLGDYLGVPTRNAGAEVSLMAHPSQCAANSMLVFSPS 156
LGDY G+PT N + + + + + + P+
Sbjct 121 SGLGDYFGIPTDNIVPTIPVTGYTTLVRSEDPSIVQPT 158
>gi|639237429|ref|WP_024568106.1| hypothetical protein [Elizabethkingia anophelis]
Length=546
Score = 134 bits (337), Expect = 7e-30, Method: Compositional matrix adjust.
Identities = 117/430 (27%), Positives = 190/430 (44%), Gaps = 44/430 (10%)
Query 318 RLLAYRFRAYESVYNAYYRD--IRNNPFI---VDGRPVY----NKWLPSMKGGADNTTYS 368
R+ F AY+ +++ YYRD + ++ F+ D R ++ N W PS+ +
Sbjct 138 RVSMLPFLAYQKIWDEYYRDENLIDSVFVDKNGDKRELFIDGINYWNPSLPY-EFRQLFD 196
Query 369 LHQCNWERDFLTTAVPNPQQGA--------NAPLVGLTVGDVVTRADDGTYSIQKQTVLV 420
+ + W D+ T+A+P Q+GA A L G+ + DG+ S T
Sbjct 197 IKKRAWHHDYFTSALPFAQKGAAVKMPLQMTADLFYNPGGNTFVKKPDGSLS---HTGFR 253
Query 421 DEDGSKYGISYKVSEDGESLVGVDYDPVSEKTPVTAINSYAELAALTTTEGSGFTIETLR 480
EDGS V DG + V+ PV NS L T GS TI LR
Sbjct 254 LEDGS-------VPADGIGHLMVETSSTGNSNPVNIDNSSNLGVDLKTASGS--TINDLR 304
Query 481 YVNAYQKFLELNMRKGFSYKQIMQGRWDIDIRFDELLMPEFIGGISRELSMRTVEQTVDQ 540
Q++LE N R G Y + + + + L PEF+GG + + V Q
Sbjct 305 RAFKLQEWLEKNARAGSRYAESILSFFGVKTSDGRLQRPEFLGGNKTPILISEVLQQSST 364
Query 541 QSTSSQGQYAEALGSKSGIAGVYGSTSNNIEVFCDEESYIIGLLTVTPVPIYTQMLSKDF 600
ST+ QG A G+ F +E Y+IGL++V P Y+Q + + F
Sbjct 365 DSTTPQGNMAGH--------GISVGKEGGFSKFFEEHGYVIGLMSVIPKTSYSQGIPRHF 416
Query 601 LYNGLLDHYQPEFDRIGFQPITYKEVCPLNLGVADTVNKANQTFGYQRPWYEYVAKYDSA 660
D++ P+F+ IG QP+ KE+ N+G D+ FGY + EY +
Sbjct 417 SKFDKFDYFWPQFEHIGEQPVYNKEIFAKNVGDYDS----GGVFGYVPRYSEYKYSPSTI 472
Query 661 HGLFRTNMKNFVMSRVF--SGLPQLGQQFLLVDPDTVNQVFSVTEYTDKIFGYVKFNATA 718
HG F+ + + + R+F S P+L + F+ V+ ++++F+V + +DK + ++ TA
Sbjct 473 HGDFKDTLYFWHLGRIFDSSAPPKLNRDFIEVNKSGLSRIFAVEDNSDKFYCHLYQKITA 532
Query 719 RLPISRVAIP 728
+ +S P
Sbjct 533 KRKMSYFGDP 542
Score = 62.8 bits (151), Expect = 6e-07, Method: Compositional matrix adjust.
Identities = 33/110 (30%), Positives = 61/110 (55%), Gaps = 3/110 (3%)
Query 19 NSFDWSHVNNLTTDFGRITPVFCELVPAKGSLRINPEFGLELMPMVFPVQTRMFARLNFF 78
++F+ S+ + +FG + P+ C+ + + INP+ L PM+ PV + +++F
Sbjct 15 STFNMSYDRKFSMNFGDLVPIHCQEIVPGDKISINPQHMTRLAPMLAPVMHEVNVFIHYF 74
Query 79 KVTLRSMWEDYSDFISNFRDDLEEPYILPGSQNFTTMLGTGSLGDYLGVP 128
V R +W+++ FI+ + L+ ++LP QN + SLGDYLG+P
Sbjct 75 FVPNRILWKNWEAFITGGQSGLDA-HMLPVVQNLP--VPKSSLGDYLGLP 121
>gi|649569140|gb|KDS75238.1| capsid family protein, partial [Parabacteroides distasonis str.
3999B T(B) 6]
Length=390
Score = 132 bits (331), Expect = 1e-29, Method: Compositional matrix adjust.
Identities = 111/396 (28%), Positives = 174/396 (44%), Gaps = 35/396 (9%)
Query 314 PANLRLLAYRFRAYESVYNAYYRDIRNNPFIVDGRPVYNKWLPSMKGGADNTTYSLHQCN 373
PA ++ A FRAY +YN YYRD + N LP +++ + LH+
Sbjct 2 PAGFKVSALPFRAYHLIYNEYYRDQNLTSELEITLDSGNYQLP-----VNSSLWQLHRRA 56
Query 374 WERDFLTTAVPNPQQGANAPLVGLTVGDVVTRADDGTYSIQKQTVLVDEDGSKYGISYKV 433
WE+D+ T+A+P Q+G + G++ +G ++ QK T D K
Sbjct 57 WEKDYFTSALPWVQRGPEVTVPINGGGEIPVEMKEG-FAAQKITTFPDR---------KP 106
Query 434 SEDGESLVGVDYDPVSEKTPVTAINSYAELAA---LTTTEGSGFTIETLRYVNAYQKFLE 490
E L V + +I A + + T+ G I +R NA Q++ E
Sbjct 107 ISGSEVLYSA--PSVLSYGQIGSIKGQALIEPDNFVVNTDQMGVNINDIRTSNALQRWFE 164
Query 491 LNMRKGFSYKQIMQGRWDIDIRFDELLMPEFIGGISRELSMRTVEQTVDQQSTSSQGQYA 550
N R G Y + + + + L P+F+GG +S+ V QT STS Q A
Sbjct 165 RNARSGSRYIEQILSHFGVRSSDARLQRPQFLGGGRTPISVSEVLQTSSTDSTSPQANMA 224
Query 551 EALGSKSGIAGVYGSTSNNIEVFCDEESYIIGLLTVTPVPIYTQMLSKDFLYNGLLDHYQ 610
G+ ++ + +E YI+G++++ P Y Q + KDF +D Y
Sbjct 225 G--------HGISAGVNHGFTRYFEEHGYIMGIMSIRPRTGYQQGVPKDFRKFDNMDFYF 276
Query 611 PEFDRIGFQPITYKEVCPLNLGVADTVNKANQTFGYQRPWYEYVAKYDSAHGLFRTNMKN 670
PEF +G Q I +E L L +D N+ TFGY + EY + HG FR NM
Sbjct 277 PEFAHLGEQEIKNEE---LYLNESDAANEG--TFGYTPRYAEYKYSQNEVHGDFRGNMAF 331
Query 671 FVMSRVFSGLPQLGQQFLLVDPDTVNQVFSVTEYTD 706
+ ++R+F P L F+ +P N+VF+ E +D
Sbjct 332 WHLNRIFKEKPNLNTTFVECNPS--NRVFATAETSD 365
>gi|649555287|gb|KDS61824.1| capsid family protein [Parabacteroides distasonis str. 3999B
T(B) 4]
gi|649560568|gb|KDS66876.1| capsid family protein [Parabacteroides distasonis str. 3999B
T(B) 4]
gi|649561020|gb|KDS67307.1| capsid family protein [Parabacteroides distasonis str. 3999B
T(B) 4]
gi|649562724|gb|KDS68908.1| capsid family protein [Parabacteroides distasonis str. 3999B
T(B) 6]
Length=541
Score = 132 bits (333), Expect = 2e-29, Method: Compositional matrix adjust.
Identities = 117/430 (27%), Positives = 186/430 (43%), Gaps = 41/430 (10%)
Query 281 SLWQF-GLNTASAVAFVEESETTYETTPFATQNKPANLRLLAYRFRAYESVYNAYYRDIR 339
SLW + GL + + + E ++ PA ++ A FRAY +YN YYRD
Sbjct 124 SLWDYLGLPSINQIG-----EAVFQVQSPNGVKAPAGFKVSALPFRAYHLIYNEYYRDQN 178
Query 340 NNPFIVDGRPVYNKWLPSMKGGADNTTYSLHQCNWERDFLTTAVPNPQQGANAPLVGLTV 399
+ N LP +++ + LH+ WE+D+ T+A+P Q+G +
Sbjct 179 LTSELEITLDSGNYQLP-----VNSSLWQLHRRAWEKDYFTSALPWVQRGPEVTVPINGG 233
Query 400 GDVVTRADDGTYSIQKQTVLVDEDGSKYGISYKVSEDGESLVGVDYDPVSEKTPVTAINS 459
G++ +G ++ QK T D K E L V + +I
Sbjct 234 GEIPVEMKEG-FAAQKITTFPDR---------KPISGSEVLYSA--PSVLSYGQIGSIKG 281
Query 460 YAELAA---LTTTEGSGFTIETLRYVNAYQKFLELNMRKGFSYKQIMQGRWDIDIRFDEL 516
A + + T+ G I +R NA Q++ E N R G Y + + + + L
Sbjct 282 QALIEPDNFVVNTDQMGVNINDIRTSNALQRWFERNARSGSRYIEQILSHFGVRSSDARL 341
Query 517 LMPEFIGGISRELSMRTVEQTVDQQSTSSQGQYAEALGSKSGIAGVYGSTSNNIEVFCDE 576
P+F+GG +S+ V QT STS Q A G+ ++ + +E
Sbjct 342 QRPQFLGGGRTPISVSEVLQTSSTDSTSPQANMAG--------HGISAGVNHGFTRYFEE 393
Query 577 ESYIIGLLTVTPVPIYTQMLSKDFLYNGLLDHYQPEFDRIGFQPITYKEVCPLNLGVADT 636
YI+G++++ P Y Q + KDF +D Y PEF +G Q I +E L L +D
Sbjct 394 HGYIMGIMSIRPRTGYQQGVPKDFRKFDNMDFYFPEFAHLGEQEIKNEE---LYLNESDA 450
Query 637 VNKANQTFGYQRPWYEYVAKYDSAHGLFRTNMKNFVMSRVFSGLPQLGQQFLLVDPDTVN 696
N+ TFGY + EY + HG FR NM + ++R+F P L F+ +P N
Sbjct 451 ANEG--TFGYTPRYAEYKYSQNEVHGDFRGNMAFWHLNRIFKEKPNLNTTFVECNPS--N 506
Query 697 QVFSVTEYTD 706
+VF+ E +D
Sbjct 507 RVFATAETSD 516
Score = 69.3 bits (168), Expect = 5e-09, Method: Compositional matrix adjust.
Identities = 45/140 (32%), Positives = 67/140 (48%), Gaps = 9/140 (6%)
Query 4 NIFDATFDANNRIDVNSFDWSHVNNLTTDFGRITPVFCELVPAKGSLRINPEFGLELMPM 63
NIF++ R N F+ S+ N LT + G + P+ C+ V R+N E + L P+
Sbjct 3 NIFNSV--KLKRPRRNVFNLSYENKLTVNAGELIPIMCKPVVPGDKFRVNTEMLVRLAPL 60
Query 64 VFPVQTRMFARLNFFKVTLRSMWEDYSDFISNFRDDLEEP----YILPG---SQNFTTML 116
V P+ R+ ++F V R +W + DFI+ D + P Y P + N
Sbjct 61 VAPMMHRVDVFTHYFFVPNRLIWNKWEDFITKGVDGTDSPVFPTYSFPSTVDTANAHNSF 120
Query 117 GTGSLGDYLGVPTRNAGAEV 136
G GSL DYLG+P+ N E
Sbjct 121 GDGSLWDYLGLPSINQIGEA 140
>gi|547920049|ref|WP_022322420.1| capsid protein VP1 [Parabacteroides merdae CAG:48]
gi|524592961|emb|CDD13573.1| capsid protein VP1 [Parabacteroides merdae CAG:48]
Length=553
Score = 130 bits (327), Expect = 1e-28, Method: Compositional matrix adjust.
Identities = 121/439 (28%), Positives = 185/439 (42%), Gaps = 39/439 (9%)
Query 278 ASSSLWQF-GLNTASAVAFVEESETTYETTPFATQNKPANLRLLAYRFRAYESVYNAYYR 336
SSLW + GL T SA +Y+ P+ ++ A FRAY+ +YN YYR
Sbjct 125 GDSSLWDYLGLPTLSACG-----NKSYDVV--NGVKVPSGFQVSALPFRAYQLIYNEYYR 177
Query 337 DIRNNPFIVDGRPVYNKWLPSMKGGADN--TTYSLHQCNWERDFLTTAVPNPQQGANAPL 394
D P+ G D SL + WE+D+ T+A+P Q+G +
Sbjct 178 DQNLT------EPIDFTLGSGTTVGGDQLMALMSLRRRAWEKDYFTSALPWLQRGPEVTV 231
Query 395 VGLTVG---DVVTRADDGTYSIQKQTVLVDEDGSKYGISYKVSEDGESLVGVDYDP-VSE 450
G DVV + + E+G Y I+ + D S + V + +
Sbjct 232 PVQGAGGSMDVVYERQSDSQKWVDSSGREFENGHAYDITMARANDPNSALMVAVNGGTNN 291
Query 451 KTPVTAINSYAELAALTTTEGSGFTIETLRYVNAYQKFLELNMRKGFSYKQIMQGRWDID 510
+ P N ++ + G I LR NA Q++ E N R G Y + + + +
Sbjct 292 RAPELDPNGTLKV----NVDEMGININDLRTSNALQRWFERNARGGSRYIEQILSHFGVR 347
Query 511 IRFDELLMPEFIGGISRELSMRTVEQTVDQQSTSSQGQYAEALGSKSGIAGVYGSTSNNI 570
L P+F+GG +S+ V QT TS Q A G+ +N
Sbjct 348 SSDARLQRPQFLGGGRMPISVSEVLQTSSTDETSPQANMAGH--------GISAGINNGF 399
Query 571 EVFCDEESYIIGLLTVTPVPIYTQMLSKDFLYNGLLDHYQPEFDRIGFQPITYKEVCPLN 630
+ + +E YIIG++++TP Y Q + +DF +D Y PEF + Q I +E
Sbjct 400 KHYFEEHGYIIGIMSITPRSGYQQGVPRDFTKFDNMDFYFPEFAHLSEQEIKNQE----- 454
Query 631 LGVADTVNKANQTFGYQRPWYEYVAKYDSAHGLFRTNMKNFVMSRVFSGLPQLGQQFLLV 690
L V++ N TFGY + EY AHG FR N+ + ++R+F P L F+
Sbjct 455 LFVSEDAAYNNGTFGYTPRYAEYKYHPSEAHGDFRGNLSFWHLNRIFEDKPNLNTTFVEC 514
Query 691 DPDTVNQVFSVTEYTDKIF 709
P N+VF+ +E D F
Sbjct 515 KPS--NRVFATSETEDDKF 531
Score = 60.1 bits (144), Expect = 4e-06, Method: Compositional matrix adjust.
Identities = 39/140 (28%), Positives = 67/140 (48%), Gaps = 13/140 (9%)
Query 4 NIFDATFDANNRIDVNSFDWSHVNNLTTDFGRITPVFCELVPAKGSLRINPEFGLELMPM 63
NIF++ R N+F+ S+ + LT + G + P+ C V + R+ E + L P+
Sbjct 3 NIFNSI--RMKRPRRNAFNLSYESKLTLNMGELVPIMCMPVVSGDKFRVKTESLVRLAPL 60
Query 64 VFPVQTRMFARLNFFKVTLRSMWEDYSDFISNFRDDLEEP-----------YILPGSQNF 112
V P+ R+ ++F V R +W ++ DFI+ D + P +++ +
Sbjct 61 VAPMMHRVNVFTHYFFVPNRLVWNEWEDFITKGVDGEDMPMFPKIQINQDSHLVSSASLI 120
Query 113 TTMLGTGSLGDYLGVPTRNA 132
G SL DYLG+PT +A
Sbjct 121 KEYFGDSSLWDYLGLPTLSA 140
>gi|609718276|emb|CDN73650.1| conserved hypothetical protein [Elizabethkingia anophelis]
Length=537
Score = 119 bits (298), Expect = 5e-25, Method: Compositional matrix adjust.
Identities = 105/417 (25%), Positives = 181/417 (43%), Gaps = 39/417 (9%)
Query 324 FRAYESVYNAYYRD----------IRNNPFIVDGRPVYNKWLPSMKGGADNTTYSLHQCN 373
F AY+ +++ +YRD NP + + + LP + + + +
Sbjct 144 FLAYQKIWDEFYRDENLIQPLFRDSNGNPVKMFNDGINDHNLPPYSKFTE--LFKMRKRA 201
Query 374 WERDFLTTAVPNPQQGANAPLVGLTVGDVVTRADDGTYSIQKQTVLVDEDGSKYGISYKV 433
W D+ T+A+P Q+G + G+V TY + QT + D G+
Sbjct 202 WHHDYFTSALPFAQKGNAVKIPIFPQGNVPL-----TYEMGSQTFIKDMAGNPAPNKDLR 256
Query 434 SEDGESLVGVDYDPVSEKTPVTAINSYAELAALTTTEGSGFTIETLRYVNAYQKFLELNM 493
S+ +L V P+S ++ L +E T+ LR Q++LE N
Sbjct 257 SDVNGNLQDVSGQPLS-------LDPSKNLKLNMASENVS-TVNDLRRAFKLQEWLEKNA 308
Query 494 RKGFSYKQIMQGRWDIDIRFDELLMPEFIGGISRELSMRTVEQTVDQQSTSSQGQYAEAL 553
R G Y + + + + L PEF+GG + + V Q ST+ QG A
Sbjct 309 RAGSRYAESILSFFGVKTSDGRLQRPEFLGGNKSPIMISEVLQQSATDSTTPQGNMA--- 365
Query 554 GSKSGIAGVYGSTSNNIEVFCDEESYIIGLLTVTPVPIYTQMLSKDFLYNGLLDHYQPEF 613
G GI G + F +E Y+IGL++V P Y+Q + + F + D++ P+F
Sbjct 366 GHGIGIGKDGGFSR-----FFEEHGYVIGLMSVIPKTSYSQGIPRHFSKSDKFDYFWPQF 420
Query 614 DRIGFQPITYKEVCPLNLGVADTVNKANQTFGYQRPWYEYVAKYDSAHGLFRTNMKNFVM 673
+ IG QP+ KE+ N+ D+ FGY + EY + HG F+ ++ + +
Sbjct 421 EHIGEQPVYNKEIFAKNIDAFDS----EAVFGYLPRYSEYKFSPSTVHGDFKDDLYFWHL 476
Query 674 SRVF--SGLPQLGQQFLLVDPDTVNQVFSVTEYTDKIFGYVKFNATARLPISRVAIP 728
R+F P L Q F+ D + ++++F+V + TDK + ++ TA+ +S P
Sbjct 477 GRIFDTDKPPVLNQSFIECDKNALSRIFAVEDDTDKFYCHLYQKITAKRKMSYFGDP 533
Score = 60.5 bits (145), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 31/110 (28%), Positives = 59/110 (54%), Gaps = 3/110 (3%)
Query 19 NSFDWSHVNNLTTDFGRITPVFCELVPAKGSLRINPEFGLELMPMVFPVQTRMFARLNFF 78
++F+ S+ + +FG + P+ C+ V + INP+ L PM+ PV + +++F
Sbjct 15 STFNMSYDRKFSMNFGDLVPIHCQEVIPGDKISINPQHMTRLAPMIAPVMHEVNVFIHYF 74
Query 79 KVTLRSMWEDYSDFISNFRDDLEEPYILPGSQNFTTMLGTGSLGDYLGVP 128
V R +W ++ FI+ L++ +++P N + GSL D+LG+P
Sbjct 75 FVPNRIIWSNWEQFITGGESGLDQ-HLMPRVGNLP--VSKGSLADHLGLP 121
>gi|557745632|ref|YP_008798242.1| major capsid protein [Marine gokushovirus]
gi|530695345|gb|AGT39902.1| major capsid protein [Marine gokushovirus]
Length=538
Score = 117 bits (294), Expect = 2e-24, Method: Compositional matrix adjust.
Identities = 121/443 (27%), Positives = 190/443 (43%), Gaps = 53/443 (12%)
Query 289 TASAVAFVEESETTYETTPFATQNKPANLRLLAYRFRAYESVYNAYYRDIR-NNPFIVDG 347
TAS E S + Y F K L A RAY V+N ++RD P +D
Sbjct 137 TASGSGEAEASLSDY----FGIPTKVGGLEFSALWHRAYTLVWNDWFRDENLQAPKTID- 191
Query 348 RPVYNKWLPSMKGGADNTTYSLHQCNWERDFLTTAVPNPQQGANAPLVGLTVGDVVTRAD 407
G D TTY+L + D+ T+A+P PQ+GA+ + L VT A+
Sbjct 192 ----------TTSGNDTTTYALLNRGKKHDYFTSALPWPQKGADV-TIPLGTSAPVTTAN 240
Query 408 DGTYSIQKQTVLVDEDGSKYGISYKVSEDGESLVGVDYDPVSEKTPVTAINSYAELAALT 467
S Q T+ G+ + S + P E T A YA+L+
Sbjct 241 S---SNQDVTIFTPNIGNTHRFLNSAS--------TNVYPGDENTD-EARRLYADLS--- 285
Query 468 TTEGSGFTIETLRYVNAYQKFLELNMRKGFSYKQIMQGRWDIDIRFDELLMPEFIGGISR 527
E + TI LR A QKFLE+ R G Y ++++ +++ L PE++GG S
Sbjct 286 --EATSATINQLRLAFATQKFLEIQARGGSRYIEVIKNHFNVTSPDARLQRPEYLGGGSS 343
Query 528 ELSMRTVEQTVDQQSTSSQGQYAEALGSKSGIAGVYGSTSNNIEVFCDEESYIIGLLTVT 587
+++ V QT +T+ QG S I S + + F E + +IG+++V
Sbjct 344 PVNISPVAQTSSTDATTPQGNL-------SAIGTTVLSGHSFTKSFT-EHTIVIGMVSVR 395
Query 588 PVPIYTQMLSKDFLYNGLLDHYQPEFDRIGFQPITYKEVCPLNLGVADTVNKANQTFGYQ 647
Y Q L++ F + D+Y P IG Q + KE+ +T TFGYQ
Sbjct 396 TDLTYQQGLNRMFSRETIYDYYWPTLSTIGEQAVKNKEIYAQGSAADET------TFGYQ 449
Query 648 RPWYEYVAKYDSAHGLFRTN----MKNFVMSRVFSGLPQLGQQFLLVDPDTVNQVFSVTE 703
+ EY K S G FR+N ++++ ++ ++ LP LG ++ V V + +V
Sbjct 450 ERYAEYRYKPSSVTGKFRSNATGTLESWHYAQEYASLPLLGDSWIQVTDTNVQRTLAVAS 509
Query 704 YTDKIFGYV-KFNATARLPISRV 725
IF + K T +P++ +
Sbjct 510 EPQFIFDSLFKLRCTRPMPVNSI 532
Score = 49.3 bits (116), Expect = 0.008, Method: Compositional matrix adjust.
Identities = 36/140 (26%), Positives = 61/140 (44%), Gaps = 14/140 (10%)
Query 16 IDVNSFDWSHVNNLTTDFGRITPVFCELVPAKGSLRINPEFGLELMPMVFPVQTRMFARL 75
I ++FD SH T + G++ P++ + + N L + P F
Sbjct 29 IQRSTFDRSHGLKTTFNAGQLVPIYVDEALPGDTFSCNLTAFSRLATPIHPTMDNAFMDT 88
Query 76 NFFKVTLRSMWEDYSDFISNFRD-DLEEPYILPGSQNFT---------TMLGTG----SL 121
+FF V +R +W+D+ +F+ + L G+ +F+ T G+G SL
Sbjct 89 HFFAVPVRLVWDDFEEFMGETKTYKAAGSDRLDGTPDFSVAAPVPPTITASGSGEAEASL 148
Query 122 GDYLGVPTRNAGAEVSLMAH 141
DY G+PT+ G E S + H
Sbjct 149 SDYFGIPTKVGGLEFSALWH 168
>gi|492501782|ref|WP_005867318.1| hypothetical protein [Parabacteroides distasonis]
gi|409230408|gb|EKN23272.1| hypothetical protein HMPREF1059_03257 [Parabacteroides distasonis
CL09T03C24]
Length=538
Score = 117 bits (294), Expect = 2e-24, Method: Compositional matrix adjust.
Identities = 118/446 (26%), Positives = 178/446 (40%), Gaps = 77/446 (17%)
Query 281 SLWQF-GLNTASA---VAFVEESETTYETTPFATQNKPANLRLLAYRFRAYESVYNAYYR 336
SLW + GL T VAF S + P ++ A FRAY+ +YN YYR
Sbjct 125 SLWDYLGLPTIGGFNNVAFPNRSPNSVM--------PPVGYQVSALPFRAYQLIYNEYYR 176
Query 337 DIRNNPFIVDGRPVYNKWLPSMKGGADNTTYSLHQCN--WERDFLTTAVPNPQQGANAPL 394
D +P+ + AD T L WE+D+ T+A+P Q+G
Sbjct 177 DQNLT------KPIEFSLNSGIVLSADEVTRLLTLRRRTWEKDYFTSALPWVQRGPEV-- 228
Query 395 VGLTVGDVVTRADDGTYSIQKQTVLVDEDGSKYGISYKVSEDGESLVGVDYDPVSEKTPV 454
TV + G ++ K ++ P + P
Sbjct 229 ----------------------TVPIQGSGGNLDVTLKNDAHADTY----RMPGTSNRPA 262
Query 455 TAINSYAELAALTTTEGS--------------GFTIETLRYVNAYQKFLELNMRKGFSYK 500
A+ T+G+ G +I LR NA Q++ E N R G Y
Sbjct 263 GAMQLVGGALIAGGTDGAYLEPDNFQVNVDELGVSINDLRTSNALQRWFERNARSGSRYI 322
Query 501 QIMQGRWDIDIRFDELLMPEFIGGISRELSMRTVEQTVDQQSTSSQGQYAEALGSKSGIA 560
+ + + + L P+F+GG +S+ V QT STS Q A
Sbjct 323 EQILSHFGVRSSDARLQRPQFLGGGRTPISVSEVLQTSATDSTSPQANMAGH-------- 374
Query 561 GVYGSTSNNIEVFCDEESYIIGLLTVTPVPIYTQMLSKDFLYNGLLDHYQPEFDRIGFQP 620
G+ ++ + + +E YIIG++++ P Y Q + KDF +D Y PEF +G Q
Sbjct 375 GISAGVNHGFKRYFEEHGYIIGIMSIRPRTGYQQGVPKDFRKFDNMDFYFPEFAHLGEQE 434
Query 621 ITYKEVCPLNLGVADTVNKANQTFGYQRPWYEYVAKYDSAHGLFRTNMKNFVMSRVFSGL 680
I +EV + T N TFGY + EY + HG FR NM + ++R+FS
Sbjct 435 IKNEEVY-----LQQTPASNNGTFGYTPRYAEYKYSMNEVHGDFRGNMAFWHLNRIFSES 489
Query 681 PQLGQQFLLVDPDTVNQVFSVTEYTD 706
P L F+ +P N+VF+ E +D
Sbjct 490 PNLNTTFVECNPS--NRVFATAETSD 513
Score = 67.0 bits (162), Expect = 3e-08, Method: Compositional matrix adjust.
Identities = 44/134 (33%), Positives = 67/134 (50%), Gaps = 10/134 (7%)
Query 4 NIFDATFDANNRIDVNSFDWSHVNNLTTDFGRITPVFCELVPAKGSLRINPEFGLELMPM 63
NIF++ R N F+ S+ N LT + G + P+ C+ V R+N E + L P+
Sbjct 3 NIFNSV--KLKRPRRNVFNLSYENKLTANAGELVPIMCKPVVPGDKFRVNTEMLVRLAPL 60
Query 64 VFPVQTRMFARLNFFKVTLRSMWEDYSDFISNFRDDLEEPY-----ILPGSQNFTT---M 115
V P+ R+ ++F V R +W + DFI+ D + P + P N T+ +
Sbjct 61 VAPMMHRVDVFTHYFFVPNRLLWNQWEDFITKGVDGTDTPVFPKIALRPDWVNPTSAAVL 120
Query 116 LGTGSLGDYLGVPT 129
L GSL DYLG+PT
Sbjct 121 LDDGSLWDYLGLPT 134
>gi|444298000|dbj|GAC77839.1| major capsid protein [uncultured marine virus]
Length=480
Score = 108 bits (269), Expect = 2e-21, Method: Compositional matrix adjust.
Identities = 117/445 (26%), Positives = 190/445 (43%), Gaps = 58/445 (13%)
Query 296 VEESETTYETTPFATQNKPANLRLLAYRFRAYESVYNAYYRDIRNNPFIVDGRPVYNKWL 355
V +ET + + + + +++ A RA+ +YN YYRD +V R + + +
Sbjct 83 VNTTETVKDLQDYLGIPRLSGVQINAMPIRAFNLIYNEYYRDQD----LVPKRELEDMTI 138
Query 356 PSMKGGADNTTYSLHQCNWERDFLTTAVPNPQQGANAPLVGLTVGDVVTRADDGTY-SIQ 414
P + W++D+ T+A P Q+G P V L +GD GT S
Sbjct 139 PLIA--------------WQKDYFTSARPWTQKG---PDVTLPLGDRAPIYGIGTTGSPA 181
Query 415 KQTVLVDEDGS---KYGISYKVSEDGESLVGVDYDP-VSEKTPVTAINSYAELAALTTTE 470
Q + V+E G +YG ++ + D DP P YA+L A T
Sbjct 182 TQNINVNETGGVNREYGAAWSSETTNAIVAEHDPDPGAGSDDP----GIYADLQAAT--- 234
Query 471 GSGFTIETLRYVNAYQKFLELNMRKGFSYKQIMQGRWDIDIRFDELLMPEFIGGISRELS 530
G TI +R A Q++ E R G Y + ++ ++ + L PE++GG + +++
Sbjct 235 --GGTINDIRRAFAIQRYQEARSRYGSRYTEYLR-YLGVNPKDARLQRPEYMGGGTTQIN 291
Query 531 MRTVEQTVDQQSTSSQGQYAEALGSKSGIAGVYGS-----TSNNIEVFCDEESYIIGLLT 585
V QT + Q S+ G+ +YG SN + +E YII +L+
Sbjct 292 FSEVLQTSPEIPGEDQV-------SQFGVGDMYGHGIAAMRSNKYRRYIEEHGYIISMLS 344
Query 586 VTPVPIYTQMLSKDFLYNGLLDHYQPEFDRIGFQPITYKEVCPLNLGVADTVNKANQTFG 645
V P +YT + + +L D+YQ E + IG Q I E+ AD +TFG
Sbjct 345 VRPKTMYTNGIHRSWLRLTKEDYYQKELEHIGQQEIMNNEI------YADE-GAGTETFG 397
Query 646 YQRPWYEYVAKYDSAHGLFRTNMKNFVMSRVFSGLPQLGQQFLLVDPDTVNQVFSVTEYT 705
Y + EY FR + + M+R F P L Q F VD D ++ + +
Sbjct 398 YNDRYSEYRETPSHVSAEFRGILNYWHMAREFEAPPVLNQSF--VDCDATKRIHN-EQTQ 454
Query 706 DKIFGYVKFNATARLPISRVAIPRL 730
D ++ ++ AR +SR A PR+
Sbjct 455 DALWIMIQHKMVARRLLSRNAAPRI 479
>gi|575096093|emb|CDL66973.1| unnamed protein product [uncultured bacterium]
Length=574
Score = 107 bits (268), Expect = 4e-21, Method: Compositional matrix adjust.
Identities = 111/427 (26%), Positives = 178/427 (42%), Gaps = 38/427 (9%)
Query 321 AYRFRAYESVYNAYYRDIRNNPFIVDGRPVYNKWLPSMKGGADNTTYSL-------HQC- 372
A FRAY ++N ++RD + N + +M G N YS + C
Sbjct 165 ALPFRAYWLIWNEWFRDENLQSSVKVSMGDTNSAVDNMGSGTGNVNYSFPSGVTSYYHCA 224
Query 373 --NWERDFLTTAVPNPQQGANAPLVGLTVGDVVTRADDGTYSIQKQTVLVDED-GSKYGI 429
D+ T+ +P PQ+G P V L +G + S+ +V + D GS Y
Sbjct 225 PRGKRYDYFTSCLPWPQKG---PGVELPLGSTANVSGQNNISLTLPSVYYNGDTGSGYSN 281
Query 430 SYKVSEDGESLVGVDYDPVSEKTPVTAINSYAELAALTT--TEGSGFTIETLRYVNAYQK 487
++ G+ L + S P + ++ L+ + + TI +LR Q+
Sbjct 282 LGQMV--GKQLSSARQETYSYIKPAGNLTLNGSMSGLSVDLSSATSITINSLRQAFMLQR 339
Query 488 FLELNMRKGFSYKQIMQGRWDIDIRFDELLMPEFIGGISRELSMRTVEQTVDQQSTSSQG 547
+ E++ R G Y + +Q + + L PE++GG S ++ V QT S QG
Sbjct 340 YYEVDARGGTRYTEKLQAHFGVTNPDSRLQRPEYLGGRSSMFNINPVAQTSSTNDISPQG 399
Query 548 QYAEALGSKSGIAGVYGSTSNNIEVFCDEESYIIGLLTVTPVPIYTQMLSKDFLYNGLLD 607
A G++G T E +IGL +V Y Q + + LD
Sbjct 400 NMAA--------YGIHGRTYRAFNKSFTEFGVVIGLCSVRADLTYQQGTERMWFRKDDLD 451
Query 608 HYQPEFDRIGFQPITYKEVCPLNLGVADTVNKANQTFGYQRPWYEYVAKYDSAHGLFRTN 667
Y PEF +G Q + +E+ + ADT FGYQ + EY K + G FR+
Sbjct 452 FYWPEFAHLGEQAVLNQEIY-VQGTSADT-----GVFGYQERYAEYRYKPNKITGQFRST 505
Query 668 MKN----FVMSRVFSGLPQLGQQFLLVDPDTVNQVFSVTEYTDKIFGYVKFNATARLPIS 723
K + +++ F LP+LG QF+ P V++V +V Y + VKF+ P+
Sbjct 506 YKQTLDVWHLAQKFDSLPKLGDQFIQDHP-PVSRVVAVPSYPHFLLD-VKFHLQCVRPLP 563
Query 724 RVAIPRL 730
+IP L
Sbjct 564 LFSIPGL 570
Lambda K H a alpha
0.318 0.133 0.393 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 5702592383325