bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: All non-redundant GenBank CDS translations+PDB+SwissProt+PIR+PRF
excluding environmental samples from WGS projects
49,011,213 sequences; 17,563,301,199 total letters
Query= Contig-3_CDS_annotation_glimmer3.pl_2_6
Length=630
Score E
Sequences producing significant alignments: (Bits) Value
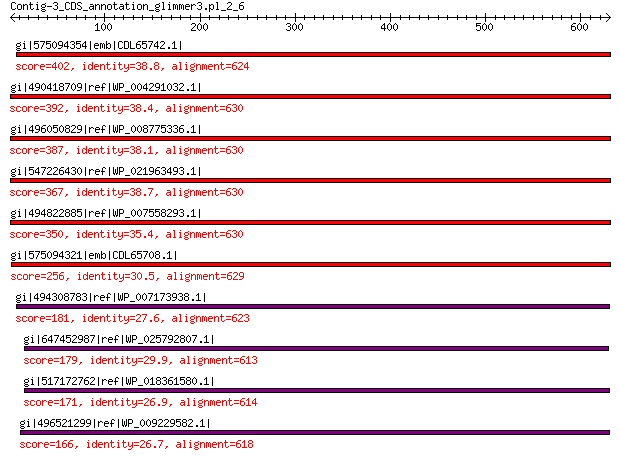
gi|575094354|emb|CDL65742.1| unnamed protein product 402 6e-128
gi|490418709|ref|WP_004291032.1| hypothetical protein 392 9e-125
gi|496050829|ref|WP_008775336.1| hypothetical protein 387 1e-122
gi|547226430|ref|WP_021963493.1| putative uncharacterized protein 367 4e-115
gi|494822885|ref|WP_007558293.1| hypothetical protein 350 6e-108
gi|575094321|emb|CDL65708.1| unnamed protein product 256 3e-72
gi|494308783|ref|WP_007173938.1| hypothetical protein 181 6e-46
gi|647452987|ref|WP_025792807.1| hypothetical protein 179 3e-45
gi|517172762|ref|WP_018361580.1| hypothetical protein 171 1e-42
gi|496521299|ref|WP_009229582.1| capsid protein 166 4e-41
>gi|575094354|emb|CDL65742.1| unnamed protein product [uncultured bacterium]
Length=615
Score = 402 bits (1033), Expect = 6e-128, Method: Compositional matrix adjust.
Identities = 242/658 (37%), Positives = 367/658 (56%), Gaps = 79/658 (12%)
Query 7 LKQLQNHPHRSGFDIGAKNVFSAKCGELLPVYWDLGIPGCTYDIDIQYFTRTRPVQTAAY 66
+ ++N P R+GFD+ K F+AK GELLPV + +PG +++I+++ FTRT+P+ T+A+
Sbjct 3 MADIKNRPSRNGFDLSFKKNFTAKAGELLPVMTKVVLPGDSFNINLRSFTRTQPLNTSAF 62
Query 67 TRIREYFDFYAVPIDLIWKSFDASVIQMGETAPVQAKDILTALT-VSGDLPYCSLSDLGL 125
R+REY+DFY VP + +W FD+ + QM + L T +SG +PY +
Sbjct 63 ARMREYYDFYFVPFEQMWNKFDSCITQMNANVQHASGPTLDDNTPLSGRMPYFT------ 116
Query 126 SCFFASGSMSVPSLKSWQANNAYANIFGYIRGDVNYKLIHMLNYGNIIPNNMPALNIGNS 185
S + + QA A N FG+ R + KL+ L YG+ N +S
Sbjct 117 -------SEQIADYLNDQATAARKNPFGFNRSTLTCKLLQYLGYGD--------YNSFDS 161
Query 186 NYRWWNREAPLASDSLIVYSQMYNFNMNVNLFPLATYQKIYQDFFRWSQWEKADPTSYNF 245
W+ + L +N+ ++ FPL YQKIY DF+R++QWEK +P+++N
Sbjct 162 ETNTWSAKPLL-------------YNLELSPFPLLAYQKIYSDFYRYTQWEKTNPSTFNL 208
Query 246 DWYQGSGNLFGGTID-TSLPASSDYWKRDNLFSLRYCNWNKDLFMGVLPNSQFGDIAVID 304
D+ +G+ +L +D T LP+ + N F +RYCN+ KD+F GVLP +Q+G +V+
Sbjct 209 DYIKGTSDL---QMDLTGLPSDDN-----NFFDIRYCNYQKDMFHGVLPVAQYGSASVVP 260
Query 305 IEGGLNIPASRIS--LSSNNRPTIGIKVGAQVSSPNNCSITNSSGNLSTGDILSVGIPA- 361
I G LN+ ++ S + + P G + V+ N + N S +S G L+VG A
Sbjct 261 INGQLNVISNGDSGPIFKTSTPDPGTPGTSYVTVGGNIGVDNRSFGVS-GSTLNVGKSAD 319
Query 362 -ASYKLQSSFN----------------------VLALRQAESLQKYREITQSVDTNYRDQ 398
+ Y S+ + +LALRQAE LQK++E++ S + +Y+ Q
Sbjct 320 PSGYGFPSNASTRSLLWENPNLIIENNQGFYVPILALRQAEFLQKWKEVSVSGEEDYKSQ 379
Query 399 IKAHFGVNVPASDSHMAQYIGGIARNLDISEVVNNNLQGDGEAVIYGKGVGTGTGSMRYT 458
I+ H+G+ V SH A+Y+GG A +LDI+EV+NNN+ GD A I GKG TG GS+R+
Sbjct 380 IEKHWGIKVSDFLSHQARYLGGCATSLDINEVINNNITGDNAADIAGKGTFTGNGSIRFE 439
Query 459 TGSKYCILMCIYHCMPVLDYDISGQHPQLLATSVDELPIPEFDNIGMEGVPLVQLVNSNL 518
+ +Y I+MCIYH +P++DY SG PIPE D IGME VPLV+ +N
Sbjct 440 SKGEYGIIMCIYHVLPIVDYVGSGVDHSCTLVDATSFPIPELDQIGMESVPLVRAMNP-- 497
Query 519 YKTNKSVKIDSILGYNPRYYAWKSNIDRIHGAFTTTLQDWVSPVDDSFLYS--TFGTPSS 576
K + + D+ LGY PRY WK+++DR G F +L+ W PV D L S + PS+
Sbjct 498 VKESDTPSADTFLGYAPRYIDWKTSVDRSVGDFADSLRTWCLPVGDKELTSANSLNFPSN 557
Query 577 GSF----VTWPFFKVNPNTLDNIFAVKSDSTWESDQFLVNSYVGCKVVRPLSRDGVPY 630
+ + FFKVNP+ +D +FAV +DST ++D+FL +S+ KVVR L +G+PY
Sbjct 558 PNVEPDSIAAGFFKVNPSIVDPLFAVVADSTVKTDEFLCSSFFDVKVVRNLDVNGLPY 615
>gi|490418709|ref|WP_004291032.1| hypothetical protein [Bacteroides eggerthii]
gi|217986636|gb|EEC52970.1| putative capsid protein (F protein) [Bacteroides eggerthii DSM
20697]
Length=578
Score = 392 bits (1008), Expect = 9e-125, Method: Compositional matrix adjust.
Identities = 242/648 (37%), Positives = 352/648 (54%), Gaps = 88/648 (14%)
Query 1 MAHFTGLKQLQNHPHRSGFDIGAKNVFSAKCGELLPVYWDLGIPGCTYDIDIQYFTRTRP 60
MA+ LK ++N P R+GFD+ K F+AK GELLPV +PG T+ I+++ FTRT+P
Sbjct 1 MANIMSLKSIRNKPSRNGFDLSFKKNFTAKAGELLPVMVKEVLPGDTFKINLKAFTRTQP 60
Query 61 VQTAAYTRIREYFDFYAVPIDLIWKSFDASVIQMGETAPVQAKDI--LTALTVSGDLPYC 118
V TAA+ RIREY+DF+ VP DL+W + + QM + P A I +SG++PY
Sbjct 61 VNTAAFARIREYYDFFFVPYDLLWNKANTVLTQMYDN-PQHAVSIDPTRNFVLSGEMPYM 119
Query 119 SLSDLGLSCFFASGSMSVPSLKSWQANNAYANIFGYIRGDVNYKLIHMLNYGN---IIPN 175
+ + S + ++ KS N FGY R + KL+ L YGN + +
Sbjct 120 TSEAIASYINALSTASALADYKS--------NYFGYNRSKSSVKLLEYLGYGNYESFLTD 171
Query 176 NMPALNIGNSNYRWWNREAPLASDSLIVYSQMYNFNMNVNLFPLATYQKIYQDFFRWSQW 235
+ WN APL + N+N N+F L YQKIY DF+R SQW
Sbjct 172 D-------------WN-TAPLMA------------NLNHNIFGLLAYQKIYSDFYRDSQW 205
Query 236 EKADPTSYNFDWYQGSGNLFGGTIDTSLPASSDYWKRDNLFSLRYCNWNKDLFMGVLPNS 295
E+ P+++N D+ GS +++ S+++++ N F LRYCNW KDLF GVLP+
Sbjct 206 ERVSPSTFNVDYLDGS------SMNLDNAYSTEFYQNYNFFDLRYCNWQKDLFHGVLPHQ 259
Query 296 QFGDIAVIDIEGGLNIPASRISLSSNNRPTIGIKVGAQVSSPNNCSITNSSGNLSTGDIL 355
Q+G+ AV I P L+ +N T+G +SP S T ++ NL D
Sbjct 260 QYGETAVASI-----TPDVTGKLTLSNFSTVG-------TSPTTASGT-ATKNLPAFD-- 304
Query 356 SVGIPAASYKLQSSFNVLALRQAESLQKYREITQSVDTNYRDQIKAHFGVNVPASDSHMA 415
+VG ++L LRQAE LQK++EITQS + +Y+DQ++ H+GV+V S +
Sbjct 305 TVG----------DLSILVLRQAEFLQKWKEITQSGNKDYKDQLEKHWGVSVGDGFSELC 354
Query 416 QYIGGIARNLDISEVVNNNLQGDGEAVIYGKGVGTGTGSMRYTTGSKYCILMCIYHCMPV 475
Y+GG++ ++DI+EV+N N+ G A I GKGVG G + + + +Y ++MCIYHC+P+
Sbjct 355 TYLGGVSSSIDINEVINTNITGSAAADIAGKGVGVANGEINFNSNGRYGLIMCIYHCLPL 414
Query 476 LDYDISGQHPQLLATSVDELPIPEFDNIGMEGVPLVQLVNSNLYKTNKSVKIDSILGYNP 535
LDY P L + + IPEFD +GM+ +PLVQL+N N S +LGY P
Sbjct 415 LDYTTDMLDPAFLKVNSTDYAIPEFDRVGMQSMPLVQLMNPLRSFANAS---GLVLGYVP 471
Query 536 RYYAWKSNIDRIHGAFTTTLQDWV-------------SPVDDSFLYSTFGTPSSGSFVTW 582
RY +K+++D+ G F TL WV P D + + PS + +
Sbjct 472 RYIDYKTSVDQSVGGFKRTLNSWVISYGNISVLKQVTLPNDAPPIEPSEPVPSVAP-MNF 530
Query 583 PFFKVNPNTLDNIFAVKSDSTWESDQFLVNSYVGCKVVRPLSRDGVPY 630
FFKVNP+ LD IFAV++ +DQFL +S+ K VR L DG+PY
Sbjct 531 TFFKVNPDCLDPIFAVQAGDDTNTDQFLCSSFFDIKAVRNLDTDGLPY 578
>gi|496050829|ref|WP_008775336.1| hypothetical protein [Bacteroides sp. 2_2_4]
gi|229448893|gb|EEO54684.1| putative capsid protein (F protein) [Bacteroides sp. 2_2_4]
Length=580
Score = 387 bits (994), Expect = 1e-122, Method: Compositional matrix adjust.
Identities = 240/637 (38%), Positives = 347/637 (54%), Gaps = 64/637 (10%)
Query 1 MAHFTGLKQLQNHPHRSGFDIGAKNVFSAKCGELLPVY-WDLGIPGCTYDIDIQYFTRTR 59
MA+ LK L+N R+GFD+ +K F+AK GELLPV W++ +PG + ID++ FTRT+
Sbjct 1 MANIMSLKSLRNKTSRNGFDLSSKRNFTAKPGELLPVKCWEV-LPGDKWSIDLKSFTRTQ 59
Query 60 PVQTAAYTRIREYFDFYAVPIDLIWKSFDASVIQMGETAPVQAKDILTA-LTVSGDLPYC 118
P+ TAA+ R+REY+DFY VP +L+W + + QM + I +A ++G +P
Sbjct 60 PLNTAAFARMREYYDFYFVPYNLLWNKANTVLTQMYDNPQHATSYIPSANQALAGVMPNV 119
Query 119 SLSDLGLSCFFASGSMSVPSLKSWQANNAYANIFGYIRGDVNYKLIHMLNYGNIIPNNMP 178
+ G++ + + V + S++ N FGY R KL+ L YGN
Sbjct 120 TCK--GIADYLNLVAPDVTTTNSYEKN-----YFGYSRSLGTAKLLEYLGYGNF------ 166
Query 179 ALNIGNSNYRWWNREAPLASDSLIVYSQMYNFNMNVNLFPLATYQKIYQDFFRWSQWEKA 238
S W + +PL+S N+ +N++ + YQKIY D R SQWEK
Sbjct 167 -YTYATSKNNTWTK-SPLSS------------NLQLNIYGVLAYQKIYADHIRDSQWEKV 212
Query 239 DPTSYNFDWYQGSGNLFGGTIDTSLPASSDYWKRDNLFSLRYCNWNKDLFMGVLPNSQFG 298
P+ +N D+ G+ + TID S+ + N+F LRYCNW KDLF GVLP Q+G
Sbjct 213 SPSCFNVDYLSGTVDS-AMTID-SMITGQGFAPFYNMFDLRYCNWQKDLFHGVLPRQQYG 270
Query 299 DIAVIDIEGGLNIPASRISLSSNNRPTIGIKVGAQVSSPNNCSITNSSGNLSTGDILSVG 358
D A +++ + A + + + P G S+ N N SG
Sbjct 271 DTAAVNVNLSNVLSAQYMVQTPDGDPVGGSPFS---STGVNLQTVNGSG----------- 316
Query 359 IPAASYKLQSSFNVLALRQAESLQKYREITQSVDTNYRDQIKAHFGVNVPASDSHMAQYI 418
+F VLALRQAE LQK++EITQS + +Y+DQI+ H+ V+V + S M+ Y+
Sbjct 317 ----------TFTVLALRQAEFLQKWKEITQSGNKDYKDQIEKHWNVSVGEAYSEMSLYL 366
Query 419 GGIARNLDISEVVNNNLQGDGEAVIYGKGVGTGTGSMRYTTGSKYCILMCIYHCMPVLDY 478
GG +LDI+EVVNNN+ G A I GKGV G G + + G +Y ++MCIYH +P+LDY
Sbjct 367 GGTTASLDINEVVNNNITGSNAADIAGKGVVVGNGRISFDAGERYGLIMCIYHSLPLLDY 426
Query 479 DISGQHPQLLATSVDELPIPEFDNIGMEGVPLVQLVNSNLYKTNKSVKIDSILGYNPRYY 538
+P + + IPEFD +GME VPLV L+N N SILGY PRY
Sbjct 427 TTDLVNPAFTKINSTDFAIPEFDRVGMESVPLVSLMNPLQSSYNVG---SSILGYAPRYI 483
Query 539 AWKSNIDRIHGAFTTTLQDWVSPVDDSFL-----YSTFGTPSSGSFVTWPFFKVNPNTLD 593
++K+++D GAF TTL+ WV D+ + Y S G+ V + FKVNPN +D
Sbjct 484 SYKTDVDSSVGAFKTTLKSWVMSYDNQSVINQLNYQDDPNNSPGTLVNYTNFKVNPNCVD 543
Query 594 NIFAVKSDSTWESDQFLVNSYVGCKVVRPLSRDGVPY 630
+FAV + ++ ++DQFL +S+ KVVR L DG+PY
Sbjct 544 PLFAVAASNSIDTDQFLCSSFFDVKVVRNLDTDGLPY 580
>gi|547226430|ref|WP_021963493.1| putative uncharacterized protein [Prevotella sp. CAG:1185]
gi|524103382|emb|CCY83994.1| putative uncharacterized protein [Prevotella sp. CAG:1185]
Length=573
Score = 367 bits (943), Expect = 4e-115, Method: Compositional matrix adjust.
Identities = 244/644 (38%), Positives = 352/644 (55%), Gaps = 85/644 (13%)
Query 1 MAHFTGLKQLQNHPHRSGFDIGAKNVFSAKCGELLPVYWDLGIPGCTYDIDIQYFTRTRP 60
M+ L L+N R+GFD+ KN F+AK GELLP+ PG ++I Q FTRT+P
Sbjct 1 MSSVMSLTALKNSVKRNGFDLSFKNAFTAKVGELLPIMCKEVYPGDKFNIRGQAFTRTQP 60
Query 61 VQTAAYTRIREYFDFYAVPIDLIWKSFDASVIQMGETAPVQAKDILTALTVSGDLPYCSL 120
V +AAY+R+REY+DFY VP L+W M + P A D+++++ +S P+ +
Sbjct 61 VNSAAYSRLREYYDFYFVPYRLLWNMAPTFFTNMPD--PHHAADLVSSVNLSQRHPWFTF 118
Query 121 SDLGLSCFFASGSMSVPSLKSWQANNAYANIFGYIRGDVNYKLIHMLNYGNIIPNNMPAL 180
D+ + S+S + + +Q N FG+ R +++ KL++ LNYG
Sbjct 119 FDI-MEYLGNLNSLS-GAYEKYQKN-----FFGFSRVELSVKLLNYLNYG---------- 161
Query 181 NIGNSNYRWWNREAPLASDSLIVYSQMYNFNMNVNLFPLATYQKIYQDFFRWSQWEKADP 240
G + + + P SD +++ + FPL YQKI +D+FR QW+ A P
Sbjct 162 -FGKD---YESVKVPSDSDDIVL-----------SPFPLLAYQKICEDYFRDDQWQSAAP 206
Query 241 TSYNFDWYQGSGNLFGGTIDTSLPASS---DYWKRDNLFSLRYCNWNKDLFMGVLPNSQF 297
YN D+ L+G + +P SS D +K +F L YCN+ KD F G+LP +Q+
Sbjct 207 YRYNLDY------LYGKSSGFHIPMSSFTNDAFKNPTMFDLNYCNFQKDYFTGMLPRAQY 260
Query 298 GDIAVID-IEGGLNIPASRISLSSNNRPTIG---IKVGAQVSSPNNCSITNSSGNLSTGD 353
GD++V I G L+I S SL+ + P G I+ G V + N +N++ LS
Sbjct 261 GDVSVASPIFGDLDIGDSS-SLTFASAPQQGANTIQSGVLVVNNN----SNTTAGLS--- 312
Query 354 ILSVGIPAASYKLQSSFNVLALRQAESLQKYREITQSVDTNYRDQIKAHFGVNVPASDSH 413
VLALRQAE LQK+REI QS +Y+ Q++ HF V+ A+ S
Sbjct 313 ------------------VLALRQAECLQKWREIAQSGKMDYQTQMQKHFNVSPSATLSG 354
Query 414 MAQYIGGIARNLDISEVVNNNLQGDGEAVIYGKGVGTGTGSMRYTTGSKYCILMCIYHCM 473
+Y+GG NLDISEVVN NL GD +A I GKG GT G+ S++ I+MCIYHC+
Sbjct 355 HCKYLGGWTSNLDISEVVNTNLTGDNQADIQGKGTGTLNGNKVDFESSEHGIIMCIYHCL 414
Query 474 PVLDYDISGQHPQLLATSVDELPIPEFDNIGMEGVPLVQLVNSNLYKTNKSVKID--SI- 530
P+LD+ I+ Q T+ + IPEFD++GM+ QL S + + + D SI
Sbjct 415 PLLDWSINRIARQNFKTTFTDYAIPEFDSVGMQ-----QLYPSEMIFGLEDLPSDPSSIN 469
Query 531 LGYNPRYYAWKSNIDRIHGAFTTTLQDWVSPVDDSFLYSTFGTPSSGSF----VTWPFFK 586
+GY PRY K++ID IHG+F TL WVSP+ DS++ + F +T+ FFK
Sbjct 470 MGYVPRYADLKTSIDEIHGSFIDTLVSWVSPLTDSYISAYRQACKDAGFSDITMTYNFFK 529
Query 587 VNPNTLDNIFAVKSDSTWESDQFLVNSYVGCKVVRPLSRDGVPY 630
VNP+ +DNIF VK+DST +DQ L+NSY K VR +G+PY
Sbjct 530 VNPHIVDNIFGVKADSTINTDQLLINSYFDIKAVRNFDYNGLPY 573
>gi|494822885|ref|WP_007558293.1| hypothetical protein [Bacteroides plebeius]
gi|198272099|gb|EDY96368.1| putative capsid protein (F protein) [Bacteroides plebeius DSM
17135]
Length=613
Score = 350 bits (897), Expect = 6e-108, Method: Compositional matrix adjust.
Identities = 223/652 (34%), Positives = 343/652 (53%), Gaps = 68/652 (10%)
Query 1 MAHFTGLKQLQNHPHRSGFDIGAKNVFSAKCGELLPVYWDLGIPGCTYDIDIQYFTRTRP 60
MA+ +K ++N P R+G+D+ K F+AK G L+PV+W +P + ++ F RT+P
Sbjct 8 MANIMSMKSVRNKPTRAGYDLTQKINFTAKAGSLIPVWWTPVLPFDDLNATVKSFVRTQP 67
Query 61 VQTAAYTRIREYFDFYAVPIDLIWKSFDASVIQM-----GETAPVQAKDILTALTVSGDL 115
+ TAA+ R+R YFDFY VP +W F ++ QM + PV A ++ +S +L
Sbjct 68 LNTAAFARMRGYFDFYFVPFRQMWNKFPTAITQMRTNLLHASGPVLADNV----PLSDEL 123
Query 116 PYCSLSDLGLSCFFASGSMSVPSLKSWQANNAYANIFGYIRGDVNYKLIHMLNYGNIIPN 175
PY + + A +S+ K N FGY R + ++ L YG+ P
Sbjct 124 PYFTAEQV------ADYIVSLADSK---------NQFGYYRAWLVCIILEYLGYGDFYPY 168
Query 176 NMPALNIGNSNYRWWNREAPLASDSLIVYSQMYNFNMNVNLFPLATYQKIYQDFFRWSQW 235
+ A G W R M N N+ + FPL YQKIY DF R++QW
Sbjct 169 IVEA--AGGEGATWATRP-------------MLN-NLKFSPFPLFAYQKIYADFNRYTQW 212
Query 236 EKADPTSYNFDWYQGSGNLFGGTIDTSLPASSDYWKRDNLFSLRYCNWNKDLFMGVLPNS 295
E+++P+++N D+ GS + +D ++ D + NLF +RY NW +DL G +P +
Sbjct 213 ERSNPSTFNIDYISGSADSL--QLDFTVEGFKDSF---NLFDMRYSNWQRDLLHGTIPQA 267
Query 296 QFGDIAVIDIEGGLNIPASRISLSSNNRPTIGIKVGAQVSSPNNCSITNSSGNL----ST 351
Q+G+ + + + G + + + P N +I SSG L S
Sbjct 268 QYGEASAVPVSGSMQV------VEGPTPPAFTTGQDGVAFLNGNVTIQGSSGYLQAQTSV 321
Query 352 GD--ILSVGIPAASYKLQ--SSFNV--LALRQAESLQKYREITQSVDTNYRDQIKAHFGV 405
G+ IL + ++ SSF V LALR+AE+ QK++E+ + + +Y QI+AH+G
Sbjct 322 GESRILRFNNTNSGLIVEGDSSFGVSILALRRAEAAQKWKEVALASEEDYPSQIEAHWGQ 381
Query 406 NVPASDSHMAQYIGGIARNLDISEVVNNNLQGDGEAVIYGKGVGTGTGSMRYTTGSKYCI 465
+V + S M Q++G I +L I+EVVNNN+ G+ A I GKG +G GS+ + G +Y I
Sbjct 382 SVNKAYSDMCQWLGSINIDLSINEVVNNNITGENAADIAGKGTMSGNGSINFNVGGQYGI 441
Query 466 LMCIYHCMPVLDYDISGQHPQLLATSVDELPIPEFDNIGMEGVPLVQLVNSNLYKTNK-S 524
+MC++H +P LDY S H T+V + PIPEFD IGME VP+++ +N K
Sbjct 442 VMCVFHVLPQLDYITSAPHFGTTLTNVLDFPIPEFDKIGMEQVPVIRGLNPVKPKDGDFK 501
Query 525 VKIDSILGYNPRYYAWKSNIDRIHGAFTTTLQDWVSPVDDSFLYS--TFGTPS----SGS 578
V + GY P+YY WK+ +D+ G F +L+ W+ P DD L + + P
Sbjct 502 VSPNLYFGYAPQYYNWKTTLDKSMGEFRRSLKTWIIPFDDEALLAADSVDFPDNPNVEAD 561
Query 579 FVTWPFFKVNPNTLDNIFAVKSDSTWESDQFLVNSYVGCKVVRPLSRDGVPY 630
V FFKV+P+ LDN+FAVK++S +DQFL ++ VVR L +G+PY
Sbjct 562 SVKAGFFKVSPSVLDNLFAVKANSDLNTDQFLCSTLFDVNVVRSLDPNGLPY 613
>gi|575094321|emb|CDL65708.1| unnamed protein product [uncultured bacterium]
Length=642
Score = 256 bits (654), Expect = 3e-72, Method: Compositional matrix adjust.
Identities = 192/663 (29%), Positives = 310/663 (47%), Gaps = 61/663 (9%)
Query 2 AHFTGLKQLQNHPHRSGFDIGAKNVFSAKCGELLPVYWDLGIPGCTYDIDIQYFTRTRPV 61
++ GL L+N P R+ FD+ +N+F+AK GELLP + PG + + YFTRT P+
Sbjct 5 SNIMGLHGLKNKPSRNSFDLSHRNMFTAKVGELLPCFVQELNPGDSVKVSSSYFTRTAPL 64
Query 62 QTAAYTRIREYFDFYAVPIDLIWKSFDASVIQMGETA-----PVQAKDILTALTVSGDLP 116
Q+ A+TR+RE ++ VP +WK FD+ V+ M + A A ++ V+ +P
Sbjct 65 QSNAFTRLRENVQYFFVPYSALWKYFDSQVLNMTKNANGGDISRIASSLVGNQKVTTQMP 124
Query 117 YCSLSDLG--LSCFFASGSMSVPSLKSWQANNAYANIFGYIRGDVNYKLIHMLNYGNIIP 174
+ L L F ++ + N G R + KL+ +L YGN P
Sbjct 125 CVNYKTLHAYLLKFINRSTVGSDGSVGPEFNR------GCYRHAESAKLLQLLGYGNF-P 177
Query 175 NNMPALNIGNSNYRWWNREAPLASDSLIVYSQMYNFNMNVNLFPLATYQKIYQDFFRWSQ 234
+ N + N+ D YN + +++F L Y KI D + + Q
Sbjct 178 EQFANFKVNNDKH---NQSGQNFKDV------TYNNSPYLSIFRLLAYHKICNDHYLYRQ 228
Query 235 WEKADPTSYNFDWYQGSGNLFGGTIDT--SLPASSDYWKRDNLFSLRYCNWNKDLFMGVL 292
W+ + + N D+ + + D S+P S ++ NL +R+ N D F GVL
Sbjct 229 WQPYNASLCNVDYLTPNSSSLLSIDDALLSIPDDSIKAEKLNLLDMRFSNLPLDYFTGVL 288
Query 293 PNSQFGDIAVIDIEGG-------LNIPASRISLSSNNRPTIG---IKVGAQVSSPNNCSI 342
P SQFG +V+++ G LN S+ S R T G ++ S+ N +
Sbjct 289 PTSQFGSESVVNLNLGNASGSAVLNGTTSKDS--GRWRTTTGEWEMEQRVASSANGNLKL 346
Query 343 TNSSGNLSTGDILSVGIPAASYKLQSSFNVLALRQAESLQKYREITQSVDTNYRDQIKAH 402
NS+G + D G A + L + +++ALR A + QKY+EI + D +++ Q++AH
Sbjct 347 DNSNGTFISHDHTFSGNVAINTSLSGNLSIIALRNALAAQKYKEIQLANDVDFQSQVEAH 406
Query 403 FGVNVPASDSHMAQYIGGIARNLDISEVVNNNLQGDGEAVIYGKGVGTGTGSMRYTTGSK 462
FG+ P + + +IGG + ++I+E +N NL GD +A G G+ S+++T +
Sbjct 407 FGIK-PDEKNENSLFIGGSSSMININEQINQNLSGDNKATYGAAPQGNGSASIKFTAKT- 464
Query 463 YCILMCIYHCMPVLDYDISGQHPQLLATSVDELPIPEFDNIGMEGVPLVQLVNSNLYKTN 522
Y +++ IY C PVLD+ G L T + IPE D+IGM+ ++ Y
Sbjct 465 YGVVIGIYRCTPVLDFAHLGIDRTLFKTDASDFVIPEMDSIGMQQTFRCEVAAPAPYNDE 524
Query 523 ---------KSVKIDSILGYNPRYYAWKSNIDRIHGAFTTTLQDWVSPVDDSFLYSTFGT 573
S + GY PRY +K++ DR +GAF +L+ WV+ ++ F
Sbjct 525 FKAFRVGDGSSPDMSETYGYAPRYSEFKTSYDRYNGAFCHSLKSWVTGIN-------FDA 577
Query 574 PSSGSFVTWP------FFKVNPNTLDNIFAVKSDSTWESDQFLVNSYVGCKVVRPLSRDG 627
+ + TW F P+ + N+F V S + + DQ V C R LSR G
Sbjct 578 IQNNVWNTWAGINAPNMFACRPDIVKNLFLVSSTNNSDDDQLYVGMVNMCYATRNLSRYG 637
Query 628 VPY 630
+PY
Sbjct 638 LPY 640
>gi|494308783|ref|WP_007173938.1| hypothetical protein [Prevotella bergensis]
gi|270333035|gb|EFA43821.1| putative capsid protein (F protein) [Prevotella bergensis DSM
17361]
Length=553
Score = 181 bits (459), Expect = 6e-46, Method: Compositional matrix adjust.
Identities = 172/639 (27%), Positives = 277/639 (43%), Gaps = 111/639 (17%)
Query 7 LKQLQNHPHRSGFDIGAKNVFSAKCGELLPVY-WDLGIPGCTYDIDIQYFTRTRPVQTAA 65
+K + + +R+ FD+ +++F+A G LLPV DL IP +I+ Q F RT P+ TAA
Sbjct 8 IKATRPNRNRNAFDLSQRHLFTAHAGMLLPVLNLDL-IPHDHVEINAQDFMRTLPMNTAA 66
Query 66 YTRIREYFDFYAVPIDLIWKSFDASVIQMGETAPVQAKDILTALTVSGDLPYCSLSDL-- 123
+ +R ++F+ VP +W FD + M + K I T +PY ++ +
Sbjct 67 FASMRGVYEFFFVPYHQLWAQFDQFITGMNDFHSSANKSIQGG-TSPLQVPYFNVDSVFN 125
Query 124 GLSCFFASGSMSVPSLKSWQANNAYA--NIFGYIRGDVNYKLIHMLNYGNIIPNNMPALN 181
L+ SGS S L+ A+ ++ GY R ++G P+N+ L
Sbjct 126 SLNTGKESGSGSTDDLQYKFKYGAFRLLDLLGYGR--------KFDSFGTAYPDNVSGLK 177
Query 182 IGNSNYRWWNREAPLASDSLIVYSQMYNFNMNVNLFPLATYQKIYQDFFRWSQWEKADPT 241
N + N ++F + Y KIYQD++R S +E D
Sbjct 178 --------------------------NNLDYNCSVFRILAYNKIYQDYYRNSNYENFDTD 211
Query 242 SYNFDWYQGSGNLFGGTIDTSLPASSDYWKRDNLFSLRYCNWNKDLFMGVLPNSQFGDIA 301
S+NFD ++G G +D + A +LF LRY N D F + + F
Sbjct 212 SFNFDKFKG------GLVDAKVVA--------DLFKLRYRNAQTDYFTNLRQSQLFSFTT 257
Query 302 VIDIEGGLNIPASRISLSSNNRPTIGIKVGAQVSSPNNCSITNSSGNLSTGDILSVGIPA 361
+ +NI A R + S+ + G S S GD
Sbjct 258 AFEDVDNINI-APRDYVKSDGSNFTRVNFGVDTDS-------------SEGD-------- 295
Query 362 ASYKLQSSFNVLALRQAESLQKYREITQSVDTNYRDQIKAHFGVNVPASDSHMAQYIGGI 421
F+V +LR A ++ K +T ++DQ++AH+GV +P S Y+GG
Sbjct 296 --------FSVSSLRAAFAVDKLLSVTMRAGKTFQDQMRAHYGVEIPDSRDGRVNYLGGF 347
Query 422 ARNLDISEVVNNN----LQGDGEAVIYGKGVGTGTGSMR---YTTGSKYCILMCIYHCMP 474
++ +S+V + + EA G+ G GTGS R ++ +LMCIY +P
Sbjct 348 DSDMQVSDVTQTSGTTATEYKPEAGYLGRVAGKGTGSGRGRIVFDAKEHGVLMCIYSLVP 407
Query 475 VLDYDISGQHPQLLATSVDELPIPEFDNIGMEGVPLVQLVNSNLYKTNKSVKIDSILGYN 534
+ YD + P + + PEF+N+GM+ PL S+ T+ + +LGY
Sbjct 408 QIQYDCTRLDPMVDKLDRFDYFTPEFENLGMQ--PLNSSYISSFCTTDPK---NPVLGYQ 462
Query 535 PRYYAWKSNIDRIHGAFTTTLQDWVSPVDDSFLYSTFGTPSSGSFVTWPFFKVNPNTLDN 594
PRY +K+ +D HG F + D +S S+ S F ++ + FK++P L++
Sbjct 463 PRYSEYKTALDVNHGQFAQS--DALS----SWSVSRFRRWTTFPQLEIADFKIDPGCLNS 516
Query 595 IFAVKSDSTWESDQFLVNSYVGCKV----VRPLSRDGVP 629
IF V + T +D Y GC V +S DG+P
Sbjct 517 IFPVDYNGTEANDCV----YGGCNFNIVKVSDMSVDGMP 551
>gi|647452987|ref|WP_025792807.1| hypothetical protein [Prevotella histicola]
Length=584
Score = 179 bits (455), Expect = 3e-45, Method: Compositional matrix adjust.
Identities = 183/650 (28%), Positives = 285/650 (44%), Gaps = 115/650 (18%)
Query 16 RSGFDIGAKNVFSAKCGELLPV-YWDLGIPGCTYDIDIQYFTRTRPVQTAAYTRIREYFD 74
R+GFD+ ++ +FSAK G+LLP+ W++ P + +Q RT + TA+Y R++EY+
Sbjct 10 RNGFDLSSRRIFSAKAGQLLPIGCWEVN-PSEHFKFSVQDLVRTTTLNTASYARMKEYYH 68
Query 75 FYAVPIDLIWKSFDASVIQMGETAPVQAKDILTALTVSGDLPYCSLSDLGLSCFFASGSM 134
F+ V +W+ FD ++ G P A L + +G Y +
Sbjct 69 FFFVSYRSLWQWFDQFIV--GTNNPHSA---LNGVKKNGTTNYNQICS------------ 111
Query 135 SVPS------LKSWQANNAYANIFGYIRGDVNYKLIHMLNYGNIIPNNMPALNIGNSNYR 188
SVP+ + + ++ + F Y G KL++MLNYG + N +N+ N
Sbjct 112 SVPTFDLGKLITRLKTSDMDSQGFNYSEGAA--KLLNMLNYG--VTNKGKFMNLEN---- 163
Query 189 WWNREAPLASDSLIVYSQMYNFNMNVNLFPLATYQKIYQDFFRWSQWEKADPTSYNFDWY 248
+ L S S +Y V+ F L YQKI+ DF+R W +D S+N D Y
Sbjct 164 LITSTSYLPSKDDKEPSSIYA--CKVSPFRLLAYQKIFNDFYRNQDWTPSDVRSFNVDDY 221
Query 249 QGSGNLFGGTIDTSLPASSDYWKRDNLFSLRYCNWNKDLFMGVLPNSQFGDIAVIDIEGG 308
NL TI+ + +RY + KD + P + D G
Sbjct 222 ADDSNL---TIEPDVALK--------FCQMRYRPYAKDWLTSMKPTPNYSD-------GI 263
Query 309 LNIPASRISLSSNNRPTIGIKVGAQVSSPNNCSITNS-SGNLSTGDILSVGIPAASYKLQ 367
N+P V N +TN+ SG++S L G + S
Sbjct 264 FNLPE-------------------YVRGNGNVILTNNKSGSVS----LDSGTVSPS---- 296
Query 368 SSFNVLALRQAESLQKYREITQSVD-TNYRDQIKAHFGVNVPASDSHMAQYIGGIARNLD 426
SF+V LR A +L K E T+ + +Y QI+AHFG VP S ++ A+++GG ++
Sbjct 297 -SFSVNDLRAAFALDKMLEATRRANGLDYASQIEAHFGFKVPESRANDARFLGGFDNSIV 355
Query 427 ISEVV--NNNLQGDGEAVIYGKGVGTGTGSMRYTT----GSKYCILMCIYHCMPVLDYDI 480
+SEVV N N DG G G G GSM T +++ I+MCIY P +Y+
Sbjct 356 VSEVVSTNGNAASDGSHASIGDLGGKGIGSMSSGTIEFDSTEHGIIMCIYSVAPQSEYNA 415
Query 481 SGQHPQLLATSVDELPIPEFDNIGMEGVPLVQLVNSNLYKTNKSVKIDSI------LGYN 534
S P + ++ PEF ++G + + L+ S L K I LGY
Sbjct 416 SYLDPFNRKLTREQFYQPEFADLGYQALIGSDLICSTLGMNEKQAGFSDIELNNNLLGYQ 475
Query 535 PRYYAWKSNIDRIHGAFTT--TLQDWVSPVDDSFLY--------------STFGTPSSGS 578
RY +K+ D + G F + +L W +P D F Y + + + S
Sbjct 476 VRYNEYKTARDLVFGDFESGKSLSYWCTPRFD-FGYGDTEKKIAPENKGGADYRKKGNRS 534
Query 579 FVTWPFFKVNPNTLDNIFAVKSDSTWESDQFLVNSYVGCKVVRPLSRDGV 628
+ F +NPN ++ IF S ++D F+VNS++ K VRP+S G+
Sbjct 535 HWSSRNFYINPNLVNPIFLT---SAVQADHFIVNSFLDVKAVRPMSVTGL 581
>gi|517172762|ref|WP_018361580.1| hypothetical protein [Prevotella nanceiensis]
Length=568
Score = 171 bits (434), Expect = 1e-42, Method: Compositional matrix adjust.
Identities = 165/634 (26%), Positives = 256/634 (40%), Gaps = 105/634 (17%)
Query 16 RSGFDIGAKNVFSAKCGELLPVYWDLGIPGCTYDIDIQYFTRTRPVQTAAYTRIREYFDF 75
R+ FDI +++F+A G LLPV +P +I+ F RT P+ +AA+ +R ++F
Sbjct 18 RNAFDISQRHLFTAPAGALLPVLSLDLLPHDHVEINASDFMRTLPMNSAAFMSMRGVYEF 77
Query 76 YAVPIDLIWKSFDASVIQMGETAPVQAKDILTALTVSGDLPYCSLSDLGLSCFFASGSMS 135
Y VP +W FD + M + Y SC S
Sbjct 78 YFVPYKQLWSGFDQFITGMSD--------------YKSSFMYAFKGKTPPSCV----SFD 119
Query 136 VPSLKSWQANNAYANIFGYIRGDVNYKLIHMLNYGNIIPNNMPALNIGNSNYRWWNREAP 195
V L W N +I G+ + Y+++ +L YG
Sbjct 120 VQKLVDWCKTNTAKDIHGFDKNKGVYRILDLLGYGK------------------------ 155
Query 196 LASDSLIVYSQMYNFNM-NVNLFPLATYQKIYQDFFRWSQWEKADPTSYNFDWYQGSGNL 254
A+ + + Y+ + M F YQKIY DF+R + +E+ S+N D + GSG
Sbjct 156 YANSAGVPYTNPTSTTMGKCTPFRGLAYQKIYNDFYRNTTYEEYQLESFNVDMFYGSGK- 214
Query 255 FGGTIDTSLPASSDYWKRDNLFSLRYCNWNKDLFMGVLPNSQFGDIAVIDIEGGLNIPAS 314
+ ++P ++ W D F+LRY N KDL V P F ++ D S
Sbjct 215 ----VKETIP--NEPWDYD-WFTLRYRNAQKDLLTNVRPTPLF---SIDDFNPQFFTGGS 264
Query 315 RISLSSNNRPTIGIKVGAQVSSPNNCSITNSSGNLSTGDILSVGIPAASYKLQSSFNVLA 374
I + T G + S+ NL + S ++ +V
Sbjct 265 DIVMEKGPNVTGG-------THEYRDSVVIVGKNLKENGVDS---------KRTMISVAD 308
Query 375 LRQAESLQKYREITQSVDTNYRDQIKAHFGVNVPASDSHMAQYIGGIARNLDISEVVNNN 434
+R A +L+K +T Y++Q++AHFG++V YIGG N+ + +V
Sbjct 309 IRNAFALEKLASVTMRAGKTYKEQMEAHFGISVEEGRDGRCTYIGGFDSNIQVGDVT--- 365
Query 435 LQGDGEAV--------------IYGKGVGTGTGSMRYTTGSKYCILMCIYHCMPVLDYDI 480
Q G V GK G+G+G +R+ ++ ILMCIY +P + YD
Sbjct 366 -QSSGTTVTGTKDTSFGGYLGRTTGKATGSGSGHIRF-DAKEHGILMCIYSLVPDVQYDS 423
Query 481 SGQHPQLLATSVDELPIPEFDNIGMEGVPLVQLVNSNLYKTNKS---VKIDSILGYNPRY 537
P + + +PEF+N+GM+ PL S Y N + +K G+ PRY
Sbjct 424 KRVDPFVQKIERGDFFVPEFENLGMQ--PLFAKNISYKYNNNTANSRIKNLGAFGWQPRY 481
Query 538 YAWKSNIDRIHGAFT--TTLQDWVSPVDDSFLYSTFGTPSSGSFVTWPFFKVNPNTLDNI 595
+K+ +D HG F L W S F + FK+NP LD++
Sbjct 482 SEYKTALDINHGQFVHQEPLSYWTVARARGESMSNFNIST---------FKINPKWLDDV 532
Query 596 FAVKSDSTWESDQFLVNSYVGCKVVRPLSRDGVP 629
FAV + T +DQ Y V +S DG+P
Sbjct 533 FAVNYNGTELTDQVFGGCYFNIVKVSDMSIDGMP 566
>gi|496521299|ref|WP_009229582.1| capsid protein [Prevotella sp. oral taxon 317]
gi|288330570|gb|EFC69154.1| putative capsid protein (F protein) [Prevotella sp. oral taxon
317 str. F0108]
Length=541
Score = 166 bits (421), Expect = 4e-41, Method: Compositional matrix adjust.
Identities = 165/641 (26%), Positives = 274/641 (43%), Gaps = 139/641 (22%)
Query 12 NHPHRSGFDIGAKNVFSAKCGELLPVYWDLGIPGCTYD---IDIQYFTRTRPVQTAAYTR 68
N P RS FD+ K++++A G LLPV L + +D I Q F RT P+ +AA+
Sbjct 15 NRP-RSAFDLSQKHLYTAPAGALLPV---LSVDLMFHDHIRIQAQDFMRTMPMNSAAFIS 70
Query 69 IREYFDFYAVPIDLIWKSFDASVIQMGETAPVQAKDILTALTVSGDLPYCSLSDLGLSCF 128
+R ++F+ VP +W +D + M + ++ + +A +GD S+ ++ L+
Sbjct 71 MRGVYEFFFVPYSQLWHPYDQFITSMND---YRSSVVSSA---AGDKALDSVPNVKLADM 124
Query 129 FASGSMSVPSLKSWQANNAYANIFGYIRGDVNYKLIHMLNYGNIIPNN---MPALNIGNS 185
+ + +IFGY + + +L+ +L YG I ++ +P L G
Sbjct 125 Y-----------KFVRERTDKDIFGYPHSNNSCRLMDLLGYGKPITSSKTPVPLLYTG-- 171
Query 186 NYRWWNREAPLASDSLIVYSQMYNFNMNVNLFPLATYQKIYQDFFRWSQWEKADPTSYNF 245
NVNLF L Y KIY D++R + +E D S+N
Sbjct 172 ---------------------------NVNLFRLLAYNKIYSDYYRNTTYEGVDVYSFNI 204
Query 246 DWYQGSGNLFGGTIDTSLPASSDYWKRDNLFSLRYCNWNKDLFMGVLPNSQFGDIAVIDI 305
D +G T +P + ++ K N L Y N D + + P F
Sbjct 205 DHKKG----------TFVPTADEFKKYLN---LHYRNAPLDFYTNLRPTPLF-------- 243
Query 306 EGGLNIPASRISLSSNNRPTIGIKVGAQVSSPNNCSITNSSGNLSTGDILSVGIPAASYK 365
++ S++ ++ Q+S P + ++ GN + ++ S +
Sbjct 244 -----------TIGSDSFSSV-----LQLSDPTGSAGFSADGNSAKLNMASPDV------ 281
Query 366 LQSSFNVLALRQAESLQKYREITQSVDTNYRDQIKAHFGVNVPASDSHMAQYIGGIARNL 425
NV A+R A +L K I+ Y +QI+AHFGV V Y+GG N+
Sbjct 282 ----LNVSAIRSAFALDKLLSISMRAGKTYAEQIEAHFGVTVSEGRDGQVYYLGGFDSNV 337
Query 426 DISEV------VNNNLQGDGEA-------VIYGKGVGTGTGSMRYTTGSKYCILMCIYHC 472
+ +V N N+ G A I GKG G+G G +++ + +LMCIY
Sbjct 338 QVGDVTQTSGTTNPNVSEVGNAKLAGYLGKITGKGTGSGYGEIQF-DAKEPGVLMCIYSV 396
Query 473 MPVLDYDISGQHPQLLATSVDELPIPEFDNIGMEGVPLV-QLVNSNLYKTNKSVKIDSIL 531
+P + YD P + + + IPEF+N+GM+ P+V V+ N K N
Sbjct 397 VPAMQYDCMRLDPFVAKQTRGDYFIPEFENLGMQ--PIVPAFVSLNRAKDNS-------Y 447
Query 532 GYNPRYYAWKSNIDRIHGAFT--TTLQDW-VSPVDDSFLYSTFGTPSSGSFVTWPFFKVN 588
G+ PRY +K+ D HG F L W ++ S +TF + K+N
Sbjct 448 GWQPRYSEYKTAFDINHGQFANGEPLSYWSIARARGSDTLNTFNVAA---------LKIN 498
Query 589 PNTLDNIFAVKSDSTWESDQFLVNSYVGCKVVRPLSRDGVP 629
P+ LD++FAV + T +D ++ + V ++ DG+P
Sbjct 499 PHWLDSVFAVNYNGTEVTDCMFGYAHFNIEKVSDMTEDGMP 539
Lambda K H a alpha
0.319 0.136 0.422 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 4767413412972