bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: All non-redundant GenBank CDS translations+PDB+SwissProt+PIR+PRF
excluding environmental samples from WGS projects
49,011,213 sequences; 17,563,301,199 total letters
Query= Contig-40_CDS_annotation_glimmer3.pl_2_4
Length=315
Score E
Sequences producing significant alignments: (Bits) Value
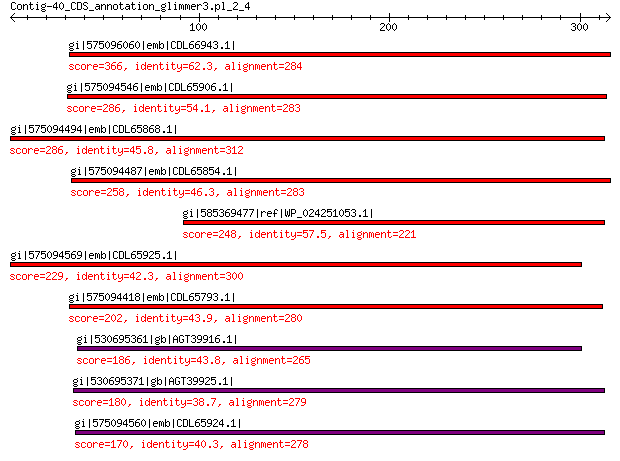
gi|575096060|emb|CDL66943.1| unnamed protein product 366 3e-122
gi|575094546|emb|CDL65906.1| unnamed protein product 286 9e-91
gi|575094494|emb|CDL65868.1| unnamed protein product 286 9e-91
gi|575094487|emb|CDL65854.1| unnamed protein product 258 5e-80
gi|585369477|ref|WP_024251053.1| hypothetical protein 248 2e-77
gi|575094569|emb|CDL65925.1| unnamed protein product 229 1e-68
gi|575094418|emb|CDL65793.1| unnamed protein product 202 5e-58
gi|530695361|gb|AGT39916.1| replication initiator 186 9e-53
gi|530695371|gb|AGT39925.1| replication initiator 180 2e-50
gi|575094560|emb|CDL65924.1| unnamed protein product 170 2e-46
>gi|575096060|emb|CDL66943.1| unnamed protein product [uncultured bacterium]
Length=339
Score = 366 bits (940), Expect = 3e-122, Method: Compositional matrix adjust.
Identities = 177/294 (60%), Positives = 218/294 (74%), Gaps = 10/294 (3%)
Query 32 EFVEIPCGKCSGCRLQRSREWANRCMLELEYHKSSYFVTLTYDDAHVPIHYYSDPETGEA 91
E V++PCG+C GCR+ SR+WANRCMLEL+ H S++F T TYD+ HVPI YY+D ETGEA
Sbjct 46 EPVQLPCGQCIGCRIDYSRQWANRCMLELQDHDSAFFCTFTYDNDHVPISYYADKETGEA 105
Query 92 LPSMSLVKRDFQLFMKRLRKKFGEG-IRFFASGEYGSLTFRPHYHAIIFGLELDDLVPYK 150
PS++L KRDFQL MKR+RK F + IRFFA+GEYG T RPHYHAII+GL L+DLVPYK
Sbjct 106 KPSLTLRKRDFQLLMKRIRKHFSDDHIRFFAAGEYGGQTLRPHYHAIIYGLHLNDLVPYK 165
Query 151 RSAQG---FQYFNSASLQEVW------PNGFAVVAPVTWETCAYTARYVMKKLTGSEAEF 201
+G + Y+NS SLQ+ W P GF VV VTWE+CAYTARYV+KK G +
Sbjct 166 TVKEGGVLYTYYNSPSLQKCWLDSDGKPIGFVVVGAVTWESCAYTARYVLKKQKGEASTV 225
Query 202 YENFNIVPEFSLMSRKPGIARQYYEDHPDLYDHEFINISTEKGGRKFRPPKYYDKLFDVD 261
Y+ FN+ PEF+LMSRKPGIAR YY+ HPDL+ +FINIST KGGRKFRPP+Y++KLF++D
Sbjct 226 YQEFNLEPEFTLMSRKPGIARNYYDTHPDLFQSDFINISTLKGGRKFRPPRYFEKLFELD 285
Query 262 CPEESARLKAVRQKMAAEAQKAKLQKTTLSYLDQLAVEERNQLARIKSLIRSCI 315
PEE+A+ VR+ + A AKL KT L L LAVEERN RIK L R+ +
Sbjct 286 FPEEAAKRSEVRKAAGSNAMAAKLAKTNLDPLSMLAVEERNFTDRIKPLRRNLV 339
>gi|575094546|emb|CDL65906.1| unnamed protein product [uncultured bacterium]
Length=351
Score = 286 bits (731), Expect = 9e-91, Method: Compositional matrix adjust.
Identities = 153/300 (51%), Positives = 193/300 (64%), Gaps = 18/300 (6%)
Query 31 TEFVEIPCGKCSGCRLQRSREWANRCMLELEYHKSSYFVTLTYDDAHVPIHYYSDPETGE 90
+ +PCG+C GCRL SR WA+R MLEL+YH ++ FVTLTY + +VP H+Y P+ G+
Sbjct 51 NNMLLLPCGQCIGCRLDYSRRWADRLMLELQYHTAAIFVTLTYSELNVPKHHYQTPD-GD 109
Query 91 ALPSMSLVKRDFQLFMKRLRKKFGEG-IRFFASGEYGSLTFRPHYHAIIFGLEL--DDLV 147
S SL KRD QLF KRLRK + + IRFF SGEYG TFRPHYHAIIFG++ D V
Sbjct 110 VNTSYSLDKRDVQLFFKRLRKMYPDTKIRFFLSGEYGPKTFRPHYHAIIFGVDFAHDRYV 169
Query 148 PYKRSAQGF--QYFNSASLQEVW-----------PNGFAVVAPVTWETCAYTARYVMKKL 194
R A Y+ S SL+ W P G + V+W TCAY ARYV KKL
Sbjct 170 WRVRRADNMFVNYYRSPSLERAWSVYNNDVGDYVPIGNVEFSDVSWHTCAYVARYVTKKL 229
Query 195 TGSEAEFYENFNIVPEFSLMSRKPGIARQYYEDHP-DLYDHEFINISTEKGGRKFRPPKY 253
TG+ A+FY FN+ P FSLMSRKPGIA QYY DH D+YD++ INISTE+GG PPKY
Sbjct 230 TGNLAQFYTTFNLTPPFSLMSRKPGIAYQYYADHGLDIYDNDKINISTERGGLSMLPPKY 289
Query 254 YDKLFDVDCPEESARLKAVRQKMAAEAQKAKLQKTTLSYLDQLAVEERNQLARIKSLIRS 313
+D +++D P++ K+VR +A E+ KL ++TLSY LAV E + +IKSL RS
Sbjct 290 FDHFYELDAPDDYVNYKSVRAALARESLLLKLDQSTLSYGQMLAVAESIKNNKIKSLDRS 349
>gi|575094494|emb|CDL65868.1| unnamed protein product [uncultured bacterium]
Length=348
Score = 286 bits (731), Expect = 9e-91, Method: Compositional matrix adjust.
Identities = 143/323 (44%), Positives = 209/323 (65%), Gaps = 12/323 (4%)
Query 1 VTSYEVDHLERSGDG---YACMRSPPTGRFGDVTEFVEIPCGKCSGCRLQRSREWANRCM 57
+ SYE DH+E DG A + V +++ IPCGKC GCRL SR+WA+RCM
Sbjct 24 ICSYESDHVELHTDGRWYVANTKDKSKYTVKVVRDYIIIPCGKCVGCRLAYSRQWADRCM 83
Query 58 LELEYHKSSYFVTLTYDDAHVPIHYYSDPETGEALPSMSLVKRDFQLFMKRLRKKFGE-- 115
LE YH SYF+TLTYDD ++P+ + +TGE + +LVK+D Q F+KRLR+ F E
Sbjct 84 LESSYHTHSYFLTLTYDDDNLPLSESINQDTGEINYNATLVKKDIQDFIKRLRR-FCEYN 142
Query 116 -----GIRFFASGEYGSLTFRPHYHAIIFGLELDDLVPYKRSAQGFQYFNSASLQEVWPN 170
I++F +GEYGS TFRPHYH I++G ++DL YK S G+ Y+NSA++ ++W
Sbjct 143 IDDNLHIKYFCAGEYGSQTFRPHYHMILYGFPINDLKLYKMSLDGYNYYNSATIDKLWKK 202
Query 171 GFAVVAPVTWETCAYTARYVMKKLTGSEAEFYENFNIVPEFSLMSRKPGIARQYYEDHPD 230
GF V+ VTW+TCAYTARY++KK GS A+ Y+++NI+PEF+ MS KP IAR+YYED+ D
Sbjct 203 GFVVIGEVTWDTCAYTARYILKKQYGSGAQIYKDYNILPEFTCMSTKPAIAREYYEDNKD 262
Query 231 -LYDHEFINISTEKGGRKFRPPKYYDKLFDVDCPEESARLKAVRQKMAAEAQKAKLQKTT 289
++D ++I + T++ + +PPKY++KL + + + + + +A + + T+
Sbjct 263 KIFDSDYIFLGTKEKSIQMKPPKYFEKLLEKENEDVFKERRDLHASLAEDFSCLRNLSTS 322
Query 290 LSYLDQLAVEERNQLARIKSLIR 312
YL L +EE N ARIK+L R
Sbjct 323 HDYLGMLQMEEDNLNARIKTLKR 345
>gi|575094487|emb|CDL65854.1| unnamed protein product [uncultured bacterium]
Length=332
Score = 258 bits (658), Expect = 5e-80, Method: Compositional matrix adjust.
Identities = 131/286 (46%), Positives = 184/286 (64%), Gaps = 4/286 (1%)
Query 33 FVEIPCGKCSGCRLQRSREWANRCMLELEYHKSSYFVTLTYDDAHVPIHYYSDPETGEAL 92
+ +PC +C GCRL +SREWANR ++E YH S+F+TLTY+D H+P + D TGE L
Sbjct 48 LLMLPCRQCVGCRLSKSREWANRVVMEQLYHVESWFLTLTYNDEHLPRSFPVDEATGEIL 107
Query 93 PSM-SLVKRDFQLFMKRLRKKFGEGIRFFASGEYGSLTFRPHYHAIIFGLELDDLVPYKR 151
+LVK D Q F+KRLRK G+ +RFFA+GEYGSL RPHYH +IFGL L+DL ++
Sbjct 108 SVHGTLVKEDLQKFLKRLRKNSGQKLRFFAAGEYGSLNMRPHYHLLIFGLHLEDLQLLRK 167
Query 152 SAQGFQYFNSASLQEVWPNGFAVVAPVTWETCAYTARYVMKKLT-GSEAEFYENFNIVPE 210
S G +Y+ S+ L++ WP GF ++ VTW++ AY ARY MKK + G + + Y+ + PE
Sbjct 168 SPLGDEYYTSSLLEKCWPFGFHILGRVTWQSAAYVARYTMKKASKGYDKDLYKKAALQPE 227
Query 211 FSLMSRKPGIARQYYEDHPDLYDHEFINISTEKGGRKFRPPKYYDKLFDVDCPEESARLK 270
F +MS +PG+ARQYYEDHPD++ + N+ST +GGRK P +Y+ KL+ D E +
Sbjct 228 FQVMSNRPGLARQYYEDHPDIFRYLSFNVSTPQGGRKMYPSEYFRKLYR-DGHERELFER 286
Query 271 AVRQKMAAEAQK-AKLQKTTLSYLDQLAVEERNQLARIKSLIRSCI 315
++R + E + K T LSY D L +E + R+ L R I
Sbjct 287 SLRTREELEVENHLKNMLTDLSYDDILKEDEEREFRRLSHLHRDLI 332
>gi|585369477|ref|WP_024251053.1| hypothetical protein [Escherichia coli]
Length=243
Score = 248 bits (633), Expect = 2e-77, Method: Compositional matrix adjust.
Identities = 127/241 (53%), Positives = 162/241 (67%), Gaps = 20/241 (8%)
Query 92 LPSMSLVKRDFQLFMKRLRKKF-GEGIRFFASGEYGSLTFRPHYHAIIFGLELDDLVPY- 149
+ S+SL KRD QLF KRLRK F + IR+FA GEYGS TFRPHYHAI+FGL L DL+P
Sbjct 1 MQSLSLCKRDLQLFWKRLRKAFPDDHIRYFACGEYGSTTFRPHYHAIVFGLHLHDLIPVQ 60
Query 150 --KRSAQGFQYFNSASLQEVW----------------PNGFAVVAPVTWETCAYTARYVM 191
+R G+QYF S SLQ W P G+ +V V WETCAY ARYV+
Sbjct 61 DIRRGDVGYQYFYSESLQRAWSVVEQKGEYDTPCIRKPIGYVLVGQVNWETCAYVARYVL 120
Query 192 KKLTGSEAEFYENFNIVPEFSLMSRKPGIARQYYEDHPDLYDHEFINISTEKGGRKFRPP 251
KK G EA+ Y+ FNI PE+ MSR+PGI RQ+Y+DHP+ +++ I+IST GGRK RPP
Sbjct 121 KKACGPEADVYQTFNIQPEYVDMSRRPGIGRQWYDDHPECMEYDTISISTPDGGRKIRPP 180
Query 252 KYYDKLFDVDCPEESARLKAVRQKMAAEAQKAKLQKTTLSYLDQLAVEERNQLARIKSLI 311
KY+DKLFD++ PE A +KA R+ A E +KAKL ++T++Y + L +ER RIK+L
Sbjct 181 KYFDKLFDLEQPELMAEIKAKRKHFAEEGKKAKLAQSTMTYEEILETQERVLHNRIKNLR 240
Query 312 R 312
R
Sbjct 241 R 241
>gi|575094569|emb|CDL65925.1| unnamed protein product [uncultured bacterium]
Length=354
Score = 229 bits (583), Expect = 1e-68, Method: Compositional matrix adjust.
Identities = 127/319 (40%), Positives = 183/319 (57%), Gaps = 23/319 (7%)
Query 1 VTSYEVDHL-----ERSGDGYACMRS------PPTGRFGDVTEFVEIPCGKCSGCRLQRS 49
+ SY+V+++ + Y C+ + P G +T+F+EIPCGKC CR + +
Sbjct 24 IVSYDVEYIVVKKYRNNKKNYVCVNTSLNEYIPVDTDVGIITDFIEIPCGKCISCRRRYA 83
Query 50 REWANRCMLELEYHKSSYFVTLTYDDAHVPIHYYSDPETGEALPSMSLVKRDFQLFMKRL 109
W +R MLEL+ HK S F+TLTYDD H+ D E + +L K Q F KRL
Sbjct 84 ALWTDRLMLELQDHKESCFITLTYDDDHICC---VDSPIEENVSMYTLNKVHLQCFWKRL 140
Query 110 RKKF------GEGIRFFASGEYGSLTFRPHYHAIIFGLELDDLVPYKRSAQGFQYFNSAS 163
R+ + IR+FA GEYG TFRPHYHAI+FG DL+ +K++ Q + S S
Sbjct 141 RQYLVRHVEPEKRIRYFACGEYGDTTFRPHYHAILFGWRPTDLIQFKKNFQNDTLYLSKS 200
Query 164 LQEVWPNGFAVVAPVTWETCAYTARYVMKKLTGSEAEFYENFNIVPEFSLMSRKPGIARQ 223
L +W NG +V VT E+C Y ARY +KK TG ++E YE ++PEF MSRKPGIAR+
Sbjct 201 LASIWQNGNVMVGDVTPESCRYVARYCLKKATGFDSEIYERLGVLPEFVTMSRKPGIARK 260
Query 224 YYEDHPD-LYDHEFINISTEKGGRKFRPPKYYDKLF-DVDCPEESARLKAVRQKMAAEAQ 281
Y++DH D + ++ IN+ST KGG + P Y+ +L D+D E +K ++ A Q
Sbjct 261 YFDDHYDEIIKYKTINLSTLKGGMSMQIPPYFIRLIEDID-SELFKEIKRSNKQAALNHQ 319
Query 282 KAKLQKTTLSYLDQLAVEE 300
+A ++ T + Y+ L+ E
Sbjct 320 EALMKNTDVDYITYLSFLE 338
>gi|575094418|emb|CDL65793.1| unnamed protein product [uncultured bacterium]
Length=367
Score = 202 bits (513), Expect = 5e-58, Method: Compositional matrix adjust.
Identities = 123/333 (37%), Positives = 180/333 (54%), Gaps = 60/333 (18%)
Query 32 EFVEIPCGKCSGCRLQRSREWANRCMLELEYHKSSYFVTLTYDDAHVPIHYYSDPETGE- 90
++V PCG+C CR+Q + WA RC LE YHK S F+TLTYD+ HVP+ + ETGE
Sbjct 40 KWVLTPCGQCLACRIQYAANWAARCELETNYHKQSIFLTLTYDEEHVPV---LNKETGEI 96
Query 91 --------------ALPSMSLVKRDFQLFMKRLRKK-----FGEGIRFFASGEYGSLTFR 131
L M++ K D Q F+KRLRK + I ++ SGEYG T R
Sbjct 97 YRGVRNPAEYVAGVTLERMTVYKPDVQKFIKRLRKAAEKEGLTDHIMYYLSGEYGDKTGR 156
Query 132 PHYHAIIFGLELDDLVPYKRSAQGFQYFNSASLQEVWPNGFAVVAPVTWETCAYTARYVM 191
PHYH I++GLE+ D + S +G+ F S L+ +W G + VT+E+C Y ARYV+
Sbjct 157 PHYHLIVYGLEVPD-AEHIGSRRGYDRFTSEWLKGIWGMGLIEIGSVTYESCQYVARYVI 215
Query 192 KKLTGSEAEFYENFNIVPEFSLMSRKPGIARQYYEDHPD-LYDHEFINISTEKGGRKFRP 250
KK G EA+ Y++ I+PEF MS KP I ++Y+E+H D +Y + IN+++ GR +P
Sbjct 216 KKRKGKEAKEYKDAGIMPEFVQMSLKPAIGQRYWEEHKDEIYSLDQINLAS---GRTVKP 272
Query 251 PKYYDKL-------------------FDVDC---PEESAR-----LKAVRQKMAAEAQKA 283
P+Y+DKL F+ DC PEE+ +K R+K +A
Sbjct 273 PRYFDKLEDQEMLMDEIGQIESLKQKFEPDCEMMPEEAESDFMRDIKKKRRKTVLAKTEA 332
Query 284 KLQKTTLSYLDQLAVEER-----NQLARIKSLI 311
+ + T L + ++ER N+LA+ + +I
Sbjct 333 RWRATNLDLREYYEMQERNFENKNKLAKNRGII 365
>gi|530695361|gb|AGT39916.1| replication initiator [Marine gokushovirus]
Length=289
Score = 186 bits (471), Expect = 9e-53, Method: Compositional matrix adjust.
Identities = 116/280 (41%), Positives = 157/280 (56%), Gaps = 41/280 (15%)
Query 36 IPCGKCSGCRLQRSREWANRCMLELEYHKSSYFVTLTYDDAHVPIHYYSDPETGEALPSM 95
+PCG+C GCRL SR+WA RC+ E + H+ + F+TLT+D+ H+ +PE
Sbjct 30 LPCGQCIGCRLDYSRQWAIRCVHEAQTHEDNCFITLTFDNEHIAKR--KNPE-------- 79
Query 96 SLVKRDFQLFMKRLRKKFGEGIRFFASGEYGSLTFRPHYHAIIFGLELDDLVPYKRSAQG 155
SL +FQ FMKRLRKK+ IRFF GEYG RPHYHA++FG + D +
Sbjct 80 SLDNTEFQRFMKRLRKKYPHKIRFFHCGEYGDQNKRPHYHALLFGHDFKDKKLWSNKGD- 138
Query 156 FQYFNSASLQEVWPNGFAVVAPVTWETCAYTARYVMKKLTGSEA---------EFYENFN 206
F+ F S L E+WP GF + V+++T AY ARYVMKK+TG A E E N
Sbjct 139 FKLFVSQELAELWPYGFHTIGAVSFDTAAYCARYVMKKVTGDAAASHYREVDLETGEVIN 198
Query 207 -IVPEFSLMSRKPGIARQYYEDHP--DLYDHEFINISTEKGGRKFRPPKYYDKLFDVDCP 263
I PE+ MSR PGI ++Y+ + D + H++I I+ G K RPP+YYDKL D
Sbjct 199 EIKPEYCTMSRMPGIGYEWYQKYGYHDCHKHDYIVIN----GYKVRPPRYYDKLCD---E 251
Query 264 EESARLKAVRQKMAAEAQKAKLQKTTLSY---LDQLAVEE 300
E A++K R A E ++Y +D+L VEE
Sbjct 252 EMFAQIKETRVANADEP--------IINYGEEMDRLWVEE 283
>gi|530695371|gb|AGT39925.1| replication initiator [Marine gokushovirus]
Length=316
Score = 180 bits (457), Expect = 2e-50, Method: Compositional matrix adjust.
Identities = 108/288 (38%), Positives = 165/288 (57%), Gaps = 31/288 (11%)
Query 34 VEIPCGKCSGCRLQRSREWANRCMLELEYHKSSYFVTLTYDDAHVPIHYYSDPETGEALP 93
++IPC +C GCRL++SR+WA RC E + +K++ F+TLTY+ H+P+ T +LP
Sbjct 47 IQIPCNQCIGCRLEKSRQWALRCTHEAKLYKNNSFITLTYNSDHLPL-------TNNSLP 99
Query 94 SMSLVKRDFQLFMKRLRKKF-GEGIRFFASGEYGSLTFRPHYHAIIFGLELDDLVPYKRS 152
+++L R FQLF+KRLRKK+ + IRF+ GEYG + RPHYHA++F + +D +K
Sbjct 100 TLNL--RHFQLFLKRLRKKYSNKTIRFYHCGEYGDMNHRPHYHALLFNHDFEDKKLWK-I 156
Query 153 AQGFQYFNSASLQEVWPN-------GFAVVAPVTWETCAYTARYVMKKLTGSEAEFYENF 205
+ Y+ S L +W + GF+ + +T+++ AY ARY +KK+TG AE Y
Sbjct 157 HKDQNYYTSEVLDGLWTDPKTKSNMGFSTIGDLTFDSAAYVARYCLKKITGKNAEDYYQ- 215
Query 206 NIVPEFSLMSRKPGIARQYYED-HPDLYDHEFINISTEKGGRKFRPPKYYDKLFDVDCPE 264
VPE++ MSR+PGI + + D+Y FI G+K +PPKYYD++ + +
Sbjct 216 GRVPEYATMSRRPGIGNGWLDKFKSDVYPSGFI----IHEGQKMQPPKYYDRVTNETDEK 271
Query 265 ESARLKAVRQKMAAEAQKAKLQKTTLSYLDQLAVEERNQLARIKSLIR 312
R K +R + EA+K T D+ V + AR K L R
Sbjct 272 AVRRSKILRMQ---EAKKHAADNTPARLSDRHTVHQ----ARAKLLKR 312
>gi|575094560|emb|CDL65924.1| unnamed protein product [uncultured bacterium]
Length=320
Score = 170 bits (430), Expect = 2e-46, Method: Compositional matrix adjust.
Identities = 112/287 (39%), Positives = 151/287 (53%), Gaps = 32/287 (11%)
Query 35 EIPCGKCSGCRLQRSREWANRCMLELEYHKSSYFVTLTYDDAHVPIHYYSDPETGEALPS 94
+IPCG+C GCRL RS + A R E + +YF+TLTY H+P P
Sbjct 44 QIPCGQCIGCRLDRSLDSAVRAHHESLLYDRNYFLTLTYSPEHLP-------------PF 90
Query 95 MSLVKRDFQLFMKRLRKKFGEGIRFFASGEYGSLTFRPHYHAIIFGLELDDLVPYKRSAQ 154
SL+ RD LF KRLRK+ G +R+ A GEYGS RPHYHAIIF L +L ++
Sbjct 91 GSLIPRDLTLFWKRLRKR-GVSLRYMACGEYGSTYGRPHYHAIIFNLPPLELKQIGTTST 149
Query 155 GFQYFNSASLQEVWPNGFAVVAPVTWETCAYTARYVMKKLTGSEAEFYENFNIVP----- 209
GF F S + E W GF + PV+++TCAY ARYV KK+ G + YE F+ V
Sbjct 150 GFPTFISDVISECWSLGFHTLNPVSFQTCAYVARYVTKKILGDGKQVYEKFDPVTGEVDC 209
Query 210 ---EFSLMSRKPGIARQYYEDH-PDLYDHEFINISTEKGGRKFRPPKYYDKLFDVDCPEE 265
EFS S KPGI Y+ + D Y + I+ +KF+ P+YYD+L D P+
Sbjct 210 RVKEFSRWSTKPGIGHDYFMKYWRDFYKIDCCLIN----NKKFKIPRYYDRLLLRDHPDV 265
Query 266 SARLKAVRQKMAAEAQKAKLQKTTLSYLDQLAVEERNQLARIKSLIR 312
+ V+QK AQ +L T + +L V E + R + L+R
Sbjct 266 ---FEIVKQKRILSAQDYRL--TPDAQKSRLLVREEVKRLRAERLLR 307
Lambda K H a alpha
0.321 0.137 0.419 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 1740253685184