bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: All non-redundant GenBank CDS translations+PDB+SwissProt+PIR+PRF
excluding environmental samples from WGS projects
49,011,213 sequences; 17,563,301,199 total letters
Query= Contig-41_CDS_annotation_glimmer3.pl_2_1
Length=85
Score E
Sequences producing significant alignments: (Bits) Value
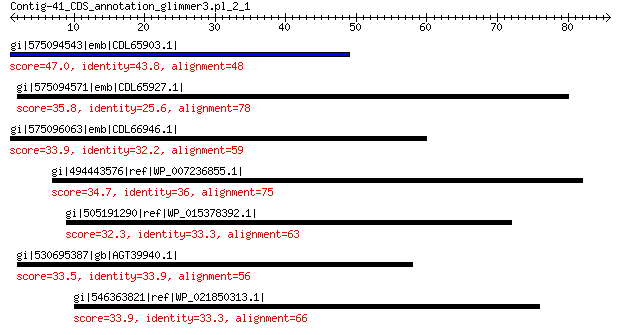
gi|575094543|emb|CDL65903.1| unnamed protein product 47.0 7e-05
gi|575094571|emb|CDL65927.1| unnamed protein product 35.8 0.75
gi|575096063|emb|CDL66946.1| unnamed protein product 33.9 5.6
gi|494443576|ref|WP_007236855.1| DNA helicase 34.7 5.8
gi|505191290|ref|WP_015378392.1| hypothetical protein 32.3 6.1
gi|530695387|gb|AGT39940.1| portal protein 33.5 7.6
gi|546363821|ref|WP_021850313.1| putative uncharacterized protein 33.9 8.2
>gi|575094543|emb|CDL65903.1| unnamed protein product [uncultured bacterium]
Length=145
Score = 47.0 bits (110), Expect = 7e-05, Method: Compositional matrix adjust.
Identities = 21/48 (44%), Positives = 30/48 (63%), Gaps = 0/48 (0%)
Query 1 MYGDFTGLPSDPIEALNLVHQSEYAFAQLSADDKAKYNNDWRRWFADL 48
+YGDFTG+P+ E LN V E AF L D++AKY + + +W + L
Sbjct 78 VYGDFTGMPASYSEILNAVLAGERAFMDLPVDERAKYGHSFAQWLSSL 125
>gi|575094571|emb|CDL65927.1| unnamed protein product [uncultured bacterium]
Length=133
Score = 35.8 bits (81), Expect = 0.75, Method: Compositional matrix adjust.
Identities = 20/78 (26%), Positives = 39/78 (50%), Gaps = 5/78 (6%)
Query 2 YGDFTGLPSDPIEALNLVHQSEYAFAQLSADDKAKYNNDWRRWFADLLSGRDNLSEKLSS 61
Y D + +P + LNL+ + E F L D +AK+++ + +W S + + +
Sbjct 55 YADLSNMPKTYADMLNLIKKGEADFLSLPVDVRAKFDHSFEKWLVTFGS-----QDWIVN 109
Query 62 VVPNSDVEKEGADSVVEK 79
+ +S V++E D + EK
Sbjct 110 MKKDSVVQEEKTDILAEK 127
>gi|575096063|emb|CDL66946.1| unnamed protein product [uncultured bacterium]
Length=163
Score = 33.9 bits (76), Expect = 5.6, Method: Compositional matrix adjust.
Identities = 19/59 (32%), Positives = 29/59 (49%), Gaps = 2/59 (3%)
Query 1 MYGDFTGLPSDPIEALNLVHQSEYAFAQLSADDKAKYNNDWRRWFADLLSGRDNLSEKL 59
+GD P EALN + + E F L D +AK+ + + + A SG D+ E+L
Sbjct 80 FFGDVLDFPQTYAEALNHMQEMERQFMSLPLDVRAKFGHSFSEFLA--ASGDDDFLERL 136
>gi|494443576|ref|WP_007236855.1| DNA helicase [Gordonia otitidis]
gi|377525044|dbj|GAB32589.1| hypothetical protein GOOTI_015_00040 [Gordonia otitidis NBRC
100426]
Length=1385
Score = 34.7 bits (78), Expect = 5.8, Method: Compositional matrix adjust.
Identities = 27/96 (28%), Positives = 42/96 (44%), Gaps = 21/96 (22%)
Query 7 GLPSDPIEALNLVHQSEYAFAQL-----SADDKAKYNNDWRR------WFADLLSGRDNL 55
GL +D + L+ VHQ + A L ++ ++K N R W +DLLS RD L
Sbjct 649 GLVADHLAVLDAVHQVQRVLADLHVPVDTSGSRSKQLNQLVRLDTQLSWVSDLLSARDQL 708
Query 56 SEKLSSVVPN----------SDVEKEGADSVVEKDS 81
+L + P ++V +EGA D+
Sbjct 709 IHELEYISPGGPRPRSVAEVAEVAREGASIAAANDA 744
>gi|505191290|ref|WP_015378392.1| hypothetical protein [Serratia marcescens]
gi|448242922|ref|YP_007406975.1| hypothetical protein SMWW4_v1c31620 [Serratia marcescens WW4]
gi|445213286|gb|AGE18956.1| hypothetical protein SMWW4_v1c31620 [Serratia marcescens WW4]
Length=63
Score = 32.3 bits (72), Expect = 6.1, Method: Compositional matrix adjust.
Identities = 21/65 (32%), Positives = 36/65 (55%), Gaps = 10/65 (15%)
Query 9 PSDPIEALNLVHQSEYAFA-QLSADDKAKYNNDWRRWFADLLSGRDNLSE-KLSSVVPNS 66
PS P ++++L+ F +LS+ +K Y N R W + +D LS+ K+S ++PN
Sbjct 6 PSQPPQSISLI-----LFGIELSSKNKKIYKNKTRHW---PYAPQDQLSQDKISKIIPNK 57
Query 67 DVEKE 71
EK+
Sbjct 58 KAEKK 62
>gi|530695387|gb|AGT39940.1| portal protein [Marine gokushovirus]
Length=164
Score = 33.5 bits (75), Expect = 7.6, Method: Compositional matrix adjust.
Identities = 19/56 (34%), Positives = 35/56 (63%), Gaps = 2/56 (4%)
Query 2 YGDFTGLPSDPIEALNLVHQSEYAFAQLSADDKAKYNNDWRRWFADLLSGRDNLSE 57
YGDF+ + +D EAL+LV ++ F + +D + K++ND +++ + +S DN E
Sbjct 67 YGDFSQV-ADYREALDLVRDAQQEFMSVPSDIRKKFDNDPGKFY-EFVSNPDNKEE 120
>gi|546363821|ref|WP_021850313.1| putative uncharacterized protein [Firmicutes bacterium CAG:791]
gi|523979001|emb|CCX64539.1| putative uncharacterized protein [Firmicutes bacterium CAG:791]
Length=215
Score = 33.9 bits (76), Expect = 8.2, Method: Compositional matrix adjust.
Identities = 22/66 (33%), Positives = 30/66 (45%), Gaps = 2/66 (3%)
Query 10 SDPIEALNLVHQSEYAFAQLSADDKAKYNNDWRRWFADLLSGRDNLSEKLSSVVPNSDVE 69
+P E VHQ E L+ D KY D W A+L+ RD K V+ + +E
Sbjct 47 CNPFEQNGWVHQFELECRNLNILDFQKY--DVLTWLAELMKHRDAADSKRYRVLSSKFIE 104
Query 70 KEGADS 75
K G D+
Sbjct 105 KYGIDT 110
Lambda K H a alpha
0.309 0.129 0.372 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 429741524859