bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: All non-redundant GenBank CDS translations+PDB+SwissProt+PIR+PRF
excluding environmental samples from WGS projects
49,011,213 sequences; 17,563,301,199 total letters
Query= Contig-47_CDS_annotation_glimmer3.pl_2_1
Length=536
Score E
Sequences producing significant alignments: (Bits) Value
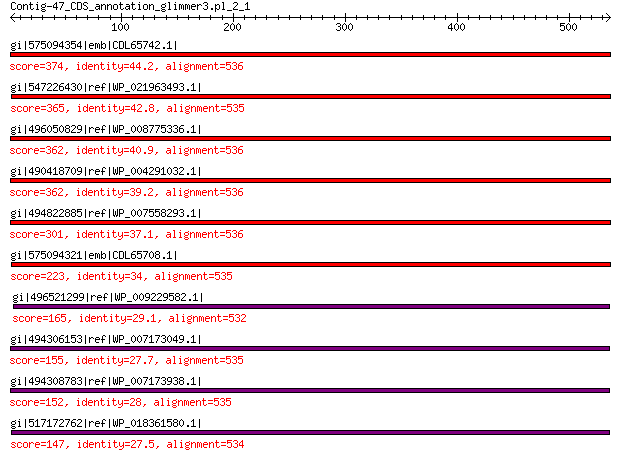
gi|575094354|emb|CDL65742.1| unnamed protein product 374 3e-118
gi|547226430|ref|WP_021963493.1| putative uncharacterized protein 365 1e-115
gi|496050829|ref|WP_008775336.1| hypothetical protein 362 3e-114
gi|490418709|ref|WP_004291032.1| hypothetical protein 362 3e-114
gi|494822885|ref|WP_007558293.1| hypothetical protein 301 1e-90
gi|575094321|emb|CDL65708.1| unnamed protein product 223 7e-61
gi|496521299|ref|WP_009229582.1| capsid protein 165 6e-41
gi|494306153|ref|WP_007173049.1| hypothetical protein 155 8e-38
gi|494308783|ref|WP_007173938.1| hypothetical protein 152 9e-37
gi|517172762|ref|WP_018361580.1| hypothetical protein 147 8e-35
>gi|575094354|emb|CDL65742.1| unnamed protein product [uncultured bacterium]
Length=615
Score = 374 bits (959), Expect = 3e-118, Method: Compositional matrix adjust.
Identities = 237/598 (40%), Positives = 331/598 (55%), Gaps = 83/598 (14%)
Query 1 MPGDKFSLSEQHFTRTQPVNTSAYTRVREYYDWFWCPLHLLWRNAPEVIAQIQQNVQHAS 60
+PGD F+++ + FTRTQP+NTSA+ R+REYYD+++ P +W I Q+ NVQHAS
Sbjct 39 LPGDSFNINLRSFTRTQPLNTSAFARMREYYDFYFVPFEQMWNKFDSCITQMNANVQHAS 98
Query 61 --SYDGSVLLGSNMPCVslsqls--kllsslkgkkNYFGFDRSDLAYKILQYLRYGNVQT 116
+ D + L MP + Q++ + +KN FGF+RS L K+LQYL YG+ +
Sbjct 99 GPTLDDNTPLSGRMPYFTSEQIADYLNDQATAARKNPFGFNRSTLTCKLLQYLGYGDYNS 158
Query 117 SSSTSGKNFGTSIPLSDRSYSQDYVFNHALSIFPLLGYKKFCQDYFRFTQWQDSAPYLWN 176
S + N ++ PL ++N LS FPLL Y+K D++R+TQW+ + P +N
Sbjct 159 FDSET--NTWSAKPL---------LYNLELSPFPLLAYQKIYSDFYRYTQWEKTNPSTFN 207
Query 177 IDYYDAKKSTSILPDTFSTSYLTHNTLIDMEYCNWNKDMFFGVLPDAQYGDASVVDI--- 233
+DY K TS L + N D+ YCN+ KDMF GVLP AQYG ASVV I
Sbjct 208 LDYI---KGTSDLQMDLTGLPSDDNNFFDIRYCNYQKDMFHGVLPVAQYGSASVVPINGQ 264
Query 234 ---------------------------------------SFGMSGQTVVASPSDISSRYT 254
SFG+SG T+ S S Y
Sbjct 265 LNVISNGDSGPIFKTSTPDPGTPGTSYVTVGGNIGVDNRSFGVSGSTLNVGKSADPSGYG 324
Query 255 ISNPSDSST-------PNL---SGSPLVLDVLALRRGEALQRFREISLCTPANYRSQIKA 304
PS++ST PNL + + +LALR+ E LQ+++E+S+ +Y+SQI+
Sbjct 325 F--PSNASTRSLLWENPNLIIENNQGFYVPILALRQAEFLQKWKEVSVSGEEDYKSQIEK 382
Query 305 HFGVDVGSELSGMSTYIGGEASSLDISEVVNTNITESNEALIAGKGIGTGQFSDKFYAK- 363
H+G+ V LS + Y+GG A+SLDI+EV+N NIT N A IAGKG TG S +F +K
Sbjct 383 HWGIKVSDFLSHQARYLGGCATSLDINEVINNNITGDNAADIAGKGTFTGNGSIRFESKG 442
Query 364 DWGILMCIYHSVPLLDYVLTSPDPQLFLSENTSFPVPELDAIGLESIPLSCYSNSSLEIP 423
++GI+MCIYH +P++DYV + D L + TSFP+PELD IG+ES+PL N P
Sbjct 443 EYGIIMCIYHVLPIVDYVGSGVDHSCTLVDATSFPIPELDQIGMESVPLVRAMN-----P 497
Query 424 ITNPNVDAASLTMGYLPRYYAWKTSLDYVLGAFTTTEKEWVAPI-TASLWSKMLLPV--- 479
+ + +A +GY PRY WKTS+D +G F + + W P+ L S L
Sbjct 498 VKESDTPSADTFLGYAPRYIDWKTSVDRSVGDFADSLRTWCLPVGDKELTSANSLNFPSN 557
Query 480 -TVDGSGINYNFFKVNPSILDPIFLVNADSTWDTDTFLVNAAFDIRVARNLDYDGMPY 536
V+ I FFKVNPSI+DP+F V ADST TD FL ++ FD++V RNLD +G+PY
Sbjct 558 PNVEPDSIAAGFFKVNPSIVDPLFAVVADSTVKTDEFLCSSFFDVKVVRNLDVNGLPY 615
>gi|547226430|ref|WP_021963493.1| putative uncharacterized protein [Prevotella sp. CAG:1185]
gi|524103382|emb|CCY83994.1| putative uncharacterized protein [Prevotella sp. CAG:1185]
Length=573
Score = 365 bits (938), Expect = 1e-115, Method: Compositional matrix adjust.
Identities = 229/559 (41%), Positives = 317/559 (57%), Gaps = 53/559 (9%)
Query 2 PGDKFSLSEQHFTRTQPVNTSAYTRVREYYDWFWCPLHLLWRNAPEVIAQIQQNVQHASS 61
PGDKF++ Q FTRTQPVN++AY+R+REYYD+++ P LLW AP + + HA+
Sbjct 44 PGDKFNIRGQAFTRTQPVNSAAYSRLREYYDFYFVPYRLLWNMAPTFFTNMP-DPHHAAD 102
Query 62 YDGSVLLGSNMPCVs--------lsqlskllsslkgkkNYFGFDRSDLAYKILQYLRYGN 113
SV L P + + S + K +KN+FGF R +L+ K+L YL YG
Sbjct 103 LVSSVNLSQRHPWFTFFDIMEYLGNLNSLSGAYEKYQKNFFGFSRVELSVKLLNYLNYG- 161
Query 114 VQTSSSTSGKNFGTSIPLSDRSYSQDYVFNHALSIFPLLGYKKFCQDYFRFTQWQDSAPY 173
GK++ + SD S D V LS FPLL Y+K C+DYFR QWQ +APY
Sbjct 162 -------FGKDYESVKVPSD---SDDIV----LSPFPLLAYQKICEDYFRDDQWQSAAPY 207
Query 174 LWNIDYYDAKKSTSILP-DTFSTSYLTHNTLIDMEYCNWNKDMFFGVLPDAQYGDASVVD 232
+N+DY K S +P +F+ + T+ D+ YCN+ KD F G+LP AQYGD SV
Sbjct 208 RYNLDYLYGKSSGFHIPMSSFTNDAFKNPTMFDLNYCNFQKDYFTGMLPRAQYGDVSVAS 267
Query 233 ISFG------MSGQTVVASPSD----ISSRYTISNPSDSSTPNLSGSPLVLDVLALRRGE 282
FG S T ++P I S + N + ++T LS VLALR+ E
Sbjct 268 PIFGDLDIGDSSSLTFASAPQQGANTIQSGVLVVNNNSNTTAGLS-------VLALRQAE 320
Query 283 ALQRFREISLCTPANYRSQIKAHFGVDVGSELSGMSTYIGGEASSLDISEVVNTNITESN 342
LQ++REI+ +Y++Q++ HF V + LSG Y+GG S+LDISEVVNTN+T N
Sbjct 321 CLQKWREIAQSGKMDYQTQMQKHFNVSPSATLSGHCKYLGGWTSNLDISEVVNTNLTGDN 380
Query 343 EALIAGKGIGT--GQFSDKFYAKDWGILMCIYHSVPLLDYVLTSPDPQLFLSENTSFPVP 400
+A I GKG GT G D F + + GI+MCIYH +PLLD+ + Q F + T + +P
Sbjct 381 QADIQGKGTGTLNGNKVD-FESSEHGIIMCIYHCLPLLDWSINRIARQNFKTTFTDYAIP 439
Query 401 ELDAIGLESIPLSCYSNSSLEIPITNPNVDAASLTMGYLPRYYAWKTSLDYVLGAFTTTE 460
E D++G++ + S ++P D +S+ MGY+PRY KTS+D + G+F T
Sbjct 440 EFDSVGMQQLYPSEMIFGLEDLP-----SDPSSINMGYVPRYADLKTSIDEIHGSFIDTL 494
Query 461 KEWVAPITASLWSKMLLPVTVDGSG---INYNFFKVNPSILDPIFLVNADSTWDTDTFLV 517
WV+P+T S S G + YNFFKVNP I+D IF V ADST +TD L+
Sbjct 495 VSWVSPLTDSYISAYRQACKDAGFSDITMTYNFFKVNPHIVDNIFGVKADSTINTDQLLI 554
Query 518 NAAFDIRVARNLDYDGMPY 536
N+ FDI+ RN DY+G+PY
Sbjct 555 NSYFDIKAVRNFDYNGLPY 573
>gi|496050829|ref|WP_008775336.1| hypothetical protein [Bacteroides sp. 2_2_4]
gi|229448893|gb|EEO54684.1| putative capsid protein (F protein) [Bacteroides sp. 2_2_4]
Length=580
Score = 362 bits (930), Expect = 3e-114, Method: Compositional matrix adjust.
Identities = 219/560 (39%), Positives = 323/560 (58%), Gaps = 46/560 (8%)
Query 1 MPGDKFSLSEQHFTRTQPVNTSAYTRVREYYDWFWCPLHLLWRNAPEVIAQIQQNVQHAS 60
+PGDK+S+ + FTRTQP+NT+A+ R+REYYD+++ P +LLW A V+ Q+ N QHA+
Sbjct 43 LPGDKWSIDLKSFTRTQPLNTAAFARMREYYDFYFVPYNLLWNKANTVLTQMYDNPQHAT 102
Query 61 SY--DGSVLLGSNMPCVslsqlsk--------llsslkgkkNYFGFDRSDLAYKILQYLR 110
SY + L MP V+ ++ + ++ +KNYFG+ RS K+L+YL
Sbjct 103 SYIPSANQALAGVMPNVTCKGIADYLNLVAPDVTTTNSYEKNYFGYSRSLGTAKLLEYLG 162
Query 111 YGNVQTSSSTSGKNFGTSIPLSDRSYSQDYVFNHALSIFPLLGYKKFCQDYFRFTQWQDS 170
YGN T + TS N T PLS N L+I+ +L Y+K D+ R +QW+
Sbjct 163 YGNFYTYA-TSKNNTWTKSPLSS---------NLQLNIYGVLAYQKIYADHIRDSQWEKV 212
Query 171 APYLWNIDYYDAKKSTSILPDTFSTS--YLTHNTLIDMEYCNWNKDMFFGVLPDAQYGDA 228
+P +N+DY +++ D+ T + + D+ YCNW KD+F GVLP QYGD
Sbjct 213 SPSCFNVDYLSGTVDSAMTIDSMITGQGFAPFYNMFDLRYCNWQKDLFHGVLPRQQYGDT 272
Query 229 SVVDISFG--MSGQTVVASPSD--ISSRYTISNPSDSSTPNLSGSPLVLDVLALRRGEAL 284
+ V+++ +S Q +V +P + S + T N SG+ VLALR+ E L
Sbjct 273 AAVNVNLSNVLSAQYMVQTPDGDPVGGSPFSSTGVNLQTVNGSGT---FTVLALRQAEFL 329
Query 285 QRFREISLCTPANYRSQIKAHFGVDVGSELSGMSTYIGGEASSLDISEVVNTNITESNEA 344
Q+++EI+ +Y+ QI+ H+ V VG S MS Y+GG +SLDI+EVVN NIT SN A
Sbjct 330 QKWKEITQSGNKDYKDQIEKHWNVSVGEAYSEMSLYLGGTTASLDINEVVNNNITGSNAA 389
Query 345 LIAGKGIGTGQFSDKFYAKD-WGILMCIYHSVPLLDYVLTSPDPQLFLSENTSFPVPELD 403
IAGKG+ G F A + +G++MCIYHS+PLLDY +P +T F +PE D
Sbjct 390 DIAGKGVVVGNGRISFDAGERYGLIMCIYHSLPLLDYTTDLVNPAFTKINSTDFAIPEFD 449
Query 404 AIGLESIPLSCYSNSSLEIPITNP---NVDAASLTMGYLPRYYAWKTSLDYVLGAFTTTE 460
+G+ES+PL + + NP + + S +GY PRY ++KT +D +GAF TT
Sbjct 450 RVGMESVPL---------VSLMNPLQSSYNVGSSILGYAPRYISYKTDVDSSVGAFKTTL 500
Query 461 KEWVAPI-TASLWSKMLL---PVTVDGSGINYNFFKVNPSILDPIFLVNADSTWDTDTFL 516
K WV S+ +++ P G+ +NY FKVNP+ +DP+F V A ++ DTD FL
Sbjct 501 KSWVMSYDNQSVINQLNYQDDPNNSPGTLVNYTNFKVNPNCVDPLFAVAASNSIDTDQFL 560
Query 517 VNAAFDIRVARNLDYDGMPY 536
++ FD++V RNLD DG+PY
Sbjct 561 CSSFFDVKVVRNLDTDGLPY 580
>gi|490418709|ref|WP_004291032.1| hypothetical protein [Bacteroides eggerthii]
gi|217986636|gb|EEC52970.1| putative capsid protein (F protein) [Bacteroides eggerthii DSM
20697]
Length=578
Score = 362 bits (929), Expect = 3e-114, Method: Compositional matrix adjust.
Identities = 210/562 (37%), Positives = 318/562 (57%), Gaps = 52/562 (9%)
Query 1 MPGDKFSLSEQHFTRTQPVNTSAYTRVREYYDWFWCPLHLLWRNAPEVIAQIQQNVQHAS 60
+PGD F ++ + FTRTQPVNT+A+ R+REYYD+F+ P LLW A V+ Q+ N QHA
Sbjct 43 LPGDTFKINLKAFTRTQPVNTAAFARIREYYDFFFVPYDLLWNKANTVLTQMYDNPQHAV 102
Query 61 SYDGS--VLLGSNMPCVslsqls-------kllsslkgkkNYFGFDRSDLAYKILQYLRY 111
S D + +L MP ++ ++ + K NYFG++RS + K+L+YL Y
Sbjct 103 SIDPTRNFVLSGEMPYMTSEAIASYINALSTASALADYKSNYFGYNRSKSSVKLLEYLGY 162
Query 112 GNVQTSSSTSGKNFGTSIPLSDRSYSQDYVFNHALSIFPLLGYKKFCQDYFRFTQWQDSA 171
GN ++ L+D + + N +IF LL Y+K D++R +QW+ +
Sbjct 163 GNYESF-------------LTDDWNTAPLMANLNHNIFGLLAYQKIYSDFYRDSQWERVS 209
Query 172 PYLWNIDYYDAKKSTSILPDTFSTSYLTHNTLIDMEYCNWNKDMFFGVLPDAQYGDASVV 231
P +N+DY D S+ L + +ST + + D+ YCNW KD+F GVLP QYG+ +V
Sbjct 210 PSTFNVDYLDG--SSMNLDNAYSTEFYQNYNFFDLRYCNWQKDLFHGVLPHQQYGETAVA 267
Query 232 DISFGMSGQTVVASPSDISSRYTISNPSDSSTPNLSGSPLV--LDVLALRRGEALQRFRE 289
I+ ++G+ +++ S + + T + S ++T NL V L +L LR+ E LQ+++E
Sbjct 268 SITPDVTGKLTLSNFSTVGTSPTTA--SGTATKNLPAFDTVGDLSILVLRQAEFLQKWKE 325
Query 290 ISLCTPANYRSQIKAHFGVDVGSELSGMSTYIGGEASSLDISEVVNTNITESNEALIAGK 349
I+ +Y+ Q++ H+GV VG S + TY+GG +SS+DI+EV+NTNIT S A IAGK
Sbjct 326 ITQSGNKDYKDQLEKHWGVSVGDGFSELCTYLGGVSSSIDINEVINTNITGSAAADIAGK 385
Query 350 GIGTGQFSDKFYAKD-WGILMCIYHSVPLLDYVLTSPDPQLFLSENTSFPVPELDAIGLE 408
G+G F + +G++MCIYH +PLLDY DP +T + +PE D +G++
Sbjct 386 GVGVANGEINFNSNGRYGLIMCIYHCLPLLDYTTDMLDPAFLKVNSTDYAIPEFDRVGMQ 445
Query 409 SIPLSCYSNSSLEIPITNP---NVDAASLTMGYLPRYYAWKTSLDYVLGAFTTTEKEWVA 465
S+PL + + NP +A+ L +GY+PRY +KTS+D +G F T WV
Sbjct 446 SMPL---------VQLMNPLRSFANASGLVLGYVPRYIDYKTSVDQSVGGFKRTLNSWVI 496
Query 466 PI-TASLWSKMLLPVTV----------DGSGINYNFFKVNPSILDPIFLVNADSTWDTDT 514
S+ ++ LP + +N+ FFKVNP LDPIF V A +TD
Sbjct 497 SYGNISVLKQVTLPNDAPPIEPSEPVPSVAPMNFTFFKVNPDCLDPIFAVQAGDDTNTDQ 556
Query 515 FLVNAAFDIRVARNLDYDGMPY 536
FL ++ FDI+ RNLD DG+PY
Sbjct 557 FLCSSFFDIKAVRNLDTDGLPY 578
>gi|494822885|ref|WP_007558293.1| hypothetical protein [Bacteroides plebeius]
gi|198272099|gb|EDY96368.1| putative capsid protein (F protein) [Bacteroides plebeius DSM
17135]
Length=613
Score = 301 bits (772), Expect = 1e-90, Method: Compositional matrix adjust.
Identities = 199/584 (34%), Positives = 296/584 (51%), Gaps = 68/584 (12%)
Query 1 MPGDKFSLSEQHFTRTQPVNTSAYTRVREYYDWFWCPLHLLWRNAPEVIAQIQQNVQHAS 60
+P D + + + F RTQP+NT+A+ R+R Y+D+++ P +W P I Q++ N+ HAS
Sbjct 50 LPFDDLNATVKSFVRTQPLNTAAFARMRGYFDFYFVPFRQMWNKFPTAITQMRTNLLHAS 109
Query 61 S--YDGSVLLGSNMPCVslsqlskllsslkgkkNYFGFDRSDLAYKILQYLRYGN----V 114
+V L +P + Q++ + SL KN FG+ R+ L IL+YL YG+ +
Sbjct 110 GPVLADNVPLSDELPYFTAEQVADYIVSLADSKNQFGYYRAWLVCIILEYLGYGDFYPYI 169
Query 115 QTSSSTSGKNFGTSIPLSDRSYSQDYVFNHALSIFPLLGYKKFCQDYFRFTQWQDSAPYL 174
++ G + T L+ N S FPL Y+K D+ R+TQW+ S P
Sbjct 170 VEAAGGEGATWATRPMLN----------NLKFSPFPLFAYQKIYADFNRYTQWERSNPST 219
Query 175 WNIDYYDAKKSTSILPDTFSTSYLTHNTLIDMEYCNWNKDMFFGVLPDAQYGDASVVDIS 234
+NIDY + S+ D + L DM Y NW +D+ G +P AQYG+AS V +S
Sbjct 220 FNIDYISGS-ADSLQLDFTVEGFKDSFNLFDMRYSNWQRDLLHGTIPQAQYGEASAVPVS 278
Query 235 FGM------------SGQTVVA-----------------SPSDISSRYTISNPSDSSTPN 265
M +GQ VA S SR N ++S
Sbjct 279 GSMQVVEGPTPPAFTTGQDGVAFLNGNVTIQGSSGYLQAQTSVGESRILRFNNTNSGLIV 338
Query 266 LSGSPLVLDVLALRRGEALQRFREISLCTPANYRSQIKAHFGVDVGSELSGMSTYIGGEA 325
S + +LALRR EA Q+++E++L + +Y SQI+AH+G V S M ++G
Sbjct 339 EGDSSFGVSILALRRAEAAQKWKEVALASEEDYPSQIEAHWGQSVNKAYSDMCQWLGSIN 398
Query 326 SSLDISEVVNTNITESNEALIAGKGIGTGQFSDKF-YAKDWGILMCIYHSVPLLDYVLTS 384
L I+EVVN NIT N A IAGKG +G S F +GI+MC++H +P LDY+ ++
Sbjct 399 IDLSINEVVNNNITGENAADIAGKGTMSGNGSINFNVGGQYGIVMCVFHVLPQLDYITSA 458
Query 385 PDPQLFLSENTSFPVPELDAIGLESIPLSCYSNSSLEIPITNPNVD---AASLTMGYLPR 441
P L+ FP+PE D IG+E +P+ N P+ + D + +L GY P+
Sbjct 459 PHFGTTLTNVLDFPIPEFDKIGMEQVPVIRGLN-----PVKPKDGDFKVSPNLYFGYAPQ 513
Query 442 YYAWKTSLDYVLGAFTTTEKEWVAPITASLWSKMLLPV---------TVDGSGINYNFFK 492
YY WKT+LD +G F + K W+ P + LL V+ + FFK
Sbjct 514 YYNWKTTLDKSMGEFRRSLKTWIIPFD----DEALLAADSVDFPDNPNVEADSVKAGFFK 569
Query 493 VNPSILDPIFLVNADSTWDTDTFLVNAAFDIRVARNLDYDGMPY 536
V+PS+LD +F V A+S +TD FL + FD+ V R+LD +G+PY
Sbjct 570 VSPSVLDNLFAVKANSDLNTDQFLCSTLFDVNVVRSLDPNGLPY 613
>gi|575094321|emb|CDL65708.1| unnamed protein product [uncultured bacterium]
Length=642
Score = 223 bits (567), Expect = 7e-61, Method: Compositional matrix adjust.
Identities = 182/614 (30%), Positives = 280/614 (46%), Gaps = 99/614 (16%)
Query 2 PGDKFSLSEQHFTRTQPVNTSAYTRVREYYDWFWCPLHLLWRNAPEVIAQIQQNVQH--- 58
PGD +S +FTRT P+ ++A+TR+RE +F+ P LW+ + + +N
Sbjct 47 PGDSVKVSSSYFTRTAPLQSNAFTRLRENVQYFFVPYSALWKYFDSQVLNMTKNANGGDI 106
Query 59 ---ASSYDGSVLLGSNMPCVslsqlskllsslkgkkNY-----------FGFDRSDLAYK 104
ASS G+ + + MPCV+ L L + G R + K
Sbjct 107 SRIASSLVGNQKVTTQMPCVNYKTLHAYLLKFINRSTVGSDGSVGPEFNRGCYRHAESAK 166
Query 105 ILQYLRYGNV----------QTSSSTSGKNFGTSIPLSDRSYSQDYVFNHA--LSIFPLL 152
+LQ L YGN + SG+NF +D +N++ LSIF LL
Sbjct 167 LLQLLGYGNFPEQFANFKVNNDKHNQSGQNF------------KDVTYNNSPYLSIFRLL 214
Query 153 GYKKFCQDYFRFTQWQDSAPYLWNIDYYDAKKST---------SILPDTFSTSYLTHNTL 203
Y K C D++ + QWQ L N+DY S+ SI D+ L L
Sbjct 215 AYHKICNDHYLYRQWQPYNASLCNVDYLTPNSSSLLSIDDALLSIPDDSIKAEKLN---L 271
Query 204 IDMEYCNWNKDMFFGVLPDAQYGDASVVDISFG-MSGQTVV-ASPSDISSRYTISNP--- 258
+DM + N D F GVLP +Q+G SVV+++ G SG V+ + S S R+ +
Sbjct 272 LDMRFSNLPLDYFTGVLPTSQFGSESVVNLNLGNASGSAVLNGTTSKDSGRWRTTTGEWE 331
Query 259 -----SDSSTPNL----------------SGSPLV-------LDVLALRRGEALQRFREI 290
+ S+ NL SG+ + L ++ALR A Q+++EI
Sbjct 332 MEQRVASSANGNLKLDNSNGTFISHDHTFSGNVAINTSLSGNLSIIALRNALAAQKYKEI 391
Query 291 SLCTPANYRSQIKAHFGVDVGSELSGMSTYIGGEASSLDISEVVNTNITESNEALIAGKG 350
L +++SQ++AHFG+ E + S +IGG +S ++I+E +N N++ N+A
Sbjct 392 QLANDVDFQSQVEAHFGIKP-DEKNENSLFIGGSSSMININEQINQNLSGDNKATYGAAP 450
Query 351 IGTGQFSDKFYAKDWGILMCIYHSVPLLDYVLTSPDPQLFLSENTSFPVPELDAIGLESI 410
G G S KF AK +G+++ IY P+LD+ D LF ++ + F +PE+D+IG++
Sbjct 451 QGNGSASIKFTAKTYGVVIGIYRCTPVLDFAHLGIDRTLFKTDASDFVIPEMDSIGMQQT 510
Query 411 PLSC-------YSNSSLEIPITNPNVDAASLTMGYLPRYYAWKTSLDYVLGAFTTTEKEW 463
C Y++ + + + S T GY PRY +KTS D GAF + K W
Sbjct 511 -FRCEVAAPAPYNDEFKAFRVGDGSSPDMSETYGYAPRYSEFKTSYDRYNGAFCHSLKSW 569
Query 464 VAPITASLWSKMLLPVTVDGSGINY-NFFKVNPSILDPIFLVNADSTWDTDTFLVNAAFD 522
V I + + V +GIN N F P I+ +FLV++ + D D V
Sbjct 570 VTGIN---FDAIQNNVWNTWAGINAPNMFACRPDIVKNLFLVSSTNNSDDDQLYVGMVNM 626
Query 523 IRVARNLDYDGMPY 536
RNL G+PY
Sbjct 627 CYATRNLSRYGLPY 640
>gi|496521299|ref|WP_009229582.1| capsid protein [Prevotella sp. oral taxon 317]
gi|288330570|gb|EFC69154.1| putative capsid protein (F protein) [Prevotella sp. oral taxon
317 str. F0108]
Length=541
Score = 165 bits (417), Expect = 6e-41, Method: Compositional matrix adjust.
Identities = 155/547 (28%), Positives = 241/547 (44%), Gaps = 70/547 (13%)
Query 4 DKFSLSEQHFTRTQPVNTSAYTRVREYYDWFWCPLHLLWRNAPEVIAQIQQ-NVQHASSY 62
D + Q F RT P+N++A+ +R Y++F+ P LW + I + SS
Sbjct 48 DHIRIQAQDFMRTMPMNSAAFISMRGVYEFFFVPYSQLWHPYDQFITSMNDYRSSVVSSA 107
Query 63 DGSVLLGSNMPCVslsqlskllsslkgkkNYFGFDRSDLAYKILQYLRYGNVQTSSSTSG 122
G L S +P V L+ + K + K + FG+ S+ + +++ L YG TSS T
Sbjct 108 AGDKALDS-VPNVKLADMYKFVRERTDK-DIFGYPHSNNSCRLMDLLGYGKPITSSKTP- 164
Query 123 KNFGTSIPLSDRSYSQDYVFNHALSIFPLLGYKKFCQDYFRFTQWQDSAPYLWNIDYYDA 182
+PL ++ +++F LL Y K DY+R T ++ Y +NID+
Sbjct 165 ------VPL---------LYTGNVNLFRLLAYNKIYSDYYRNTTYEGVDVYSFNIDH--- 206
Query 183 KKSTSI-LPDTFSTSYLTHNTLIDMEYCNWNKDMFFGVLPDAQYGDASVVDISFGMSGQT 241
KK T + D F +++ Y N D + + P + + G +
Sbjct 207 KKGTFVPTADEFKK-------YLNLHYRNAPLDFYTNLRPTPLF--------TIGSDSFS 251
Query 242 VVASPSDISSRYTISNPSDSSTPNLSGSPLVLDVLALRRGEALQRFREISLCTPANYRSQ 301
V SD + S +S+ N++ SP VL+V A+R AL + IS+ Y Q
Sbjct 252 SVLQLSDPTGSAGFSADGNSAKLNMA-SPDVLNVSAIRSAFALDKLLSISMRAGKTYAEQ 310
Query 302 IKAHFGVDVGSELSGMSTYIGGEASSLDISEVVNT------NITESNEALIA-------G 348
I+AHFGV V G Y+GG S++ + +V T N++E A +A G
Sbjct 311 IEAHFGVTVSEGRDGQVYYLGGFDSNVQVGDVTQTSGTTNPNVSEVGNAKLAGYLGKITG 370
Query 349 KGIGTGQFSDKFYAKDWGILMCIYHSVPLLDYVLTSPDPQLFLSENTSFPVPELDAIGLE 408
KG G+G +F AK+ G+LMCIY VP + Y DP + + +PE + +G++
Sbjct 371 KGTGSGYGEIQFDAKEPGVLMCIYSVVPAMQYDCMRLDPFVAKQTRGDYFIPEFENLGMQ 430
Query 409 SIPLSCYSNSSLEIPITNPNVDAASLTMGYLPRYYAWKTSLDYVLGAFTTTEKEWVAPIT 468
I +P A + G+ PRY +KT+ D G F E P+
Sbjct 431 PI-----------VPAFVSLNRAKDNSYGWQPRYSEYKTAFDINHGQFANGE-----PL- 473
Query 469 ASLWSKMLLPVTVDGSGINYNFFKVNPSILDPIFLVNADSTWDTDTFLVNAAFDIRVARN 528
S WS + + N K+NP LD +F VN + T TD A F+I +
Sbjct 474 -SYWSIARARGSDTLNTFNVAALKINPHWLDSVFAVNYNGTEVTDCMFGYAHFNIEKVSD 532
Query 529 LDYDGMP 535
+ DGMP
Sbjct 533 MTEDGMP 539
>gi|494306153|ref|WP_007173049.1| hypothetical protein [Prevotella bergensis]
gi|270333881|gb|EFA44667.1| putative capsid protein (F protein) [Prevotella bergensis DSM
17361]
Length=519
Score = 155 bits (392), Expect = 8e-38, Method: Compositional matrix adjust.
Identities = 148/556 (27%), Positives = 244/556 (44%), Gaps = 70/556 (13%)
Query 1 MPGDKFSLSEQHFTRTQPVNTSAYTRVREYYDWFWCPLHLLWRNAPEVIAQIQQNVQHAS 60
+P D ++ Q F RT P+NT+A+ +R Y++F+ P H LW + I + N H+S
Sbjct 11 IPHDHVEINAQDFMRTLPMNTAAFASMRGVYEFFFVPYHQLWAQFDQFITGM--NDFHSS 68
Query 61 SYDGSVLLGS--------NMPCVslsqlskllsslkgkkNYFGFDRSDLAYKILQYLRYG 112
+ + S+ G+ N+ V + + + + + F A+++L L YG
Sbjct 69 A-NKSIQGGTSPLQVPYFNLESVFKNIIERDSTPSFQDDLQYRFKYG--AFRLLDLLGYG 125
Query 113 NVQTSSSTSGKNFGTSIPLSDRSYSQDYVFNHALSIFPLLGYKKFCQDYFRFTQWQDSAP 172
S FGT+ P + + +N S+F +L Y K QDY+R + +++
Sbjct 126 RKFDS-------FGTAYPDNVSGLKNNLDYN--CSVFRVLAYNKIYQDYYRNSNYENFDT 176
Query 173 YLWNIDYY-----DAKKSTSILPDTFSTSYLTHNTLIDMEYCNWNKDMFFGVLPDAQYGD 227
+N D + DAK ++ D F Y N D + N + F +P ++ D
Sbjct 177 DSFNFDKFKGGLVDAK----VVADLFKLRY--RNAQTDY-FTNLRQSQLFTFIP--EFSD 227
Query 228 ASVVDISFGMSGQTVVASPSDISSRYTISNPSDSSTPNLSGSPLVLDVLALRRGEALQRF 287
++ Q S S+ + ++ P D NL V +LR A+ +
Sbjct 228 DEHLNFD---RDQYADQSKSNFTQ---LNFPVDVDN-NLG----YFSVSSLRSAFAVDKL 276
Query 288 REISLCTPANYRSQIKAHFGVDVGSELSGMSTYIGGEASSLDISEVVNTNITESNE---- 343
+++ ++ Q++AH+GV++ G Y+GG S L +S+V T+ T + E
Sbjct 277 LSVTMRAGKTFQDQMRAHYGVEIPDSRDGRVNYLGGFDSDLQVSDVTQTSGTTATEYKPE 336
Query 344 ----ALIAGKGIGTGQFSDKFYAKDWGILMCIYHSVPLLDYVLTSPDPQLFLSENTSFPV 399
IAGKG G+G+ F AK+ G+LMCIY VP + Y T DP + + F
Sbjct 337 AGYLGRIAGKGTGSGRGRIVFDAKEHGVLMCIYSLVPQIQYDCTRLDPMVDKLDRFDFFT 396
Query 400 PELDAIGLESIPLSCYSNSSLEIPITNPNVDAASLTMGYLPRYYAWKTSLDYVLGAFTTT 459
PE + +G++ PL+ SS P D + +GY PRY +KT+LD G F
Sbjct 397 PEFENLGMQ--PLNSSYISSFCTP------DPKNPVLGYQPRYSEYKTALDINHGQFAQN 448
Query 460 EKEWVAPITASLWSKMLLPVTVDGSGINYNFFKVNPSILDPIFLVNADSTWDTDTFLVNA 519
+ S WS + FK++P L+ +F V + T TD
Sbjct 449 D-------ALSSWSVSRFRRWTTFPQLEIADFKIDPGCLNSVFPVEFNGTESTDCVFGGC 501
Query 520 AFDIRVARNLDYDGMP 535
F+I ++ DGMP
Sbjct 502 NFNIVKVSDMSVDGMP 517
>gi|494308783|ref|WP_007173938.1| hypothetical protein [Prevotella bergensis]
gi|270333035|gb|EFA43821.1| putative capsid protein (F protein) [Prevotella bergensis DSM
17361]
Length=553
Score = 152 bits (385), Expect = 9e-37, Method: Compositional matrix adjust.
Identities = 150/556 (27%), Positives = 244/556 (44%), Gaps = 69/556 (12%)
Query 1 MPGDKFSLSEQHFTRTQPVNTSAYTRVREYYDWFWCPLHLLWRNAPEVIAQIQQNVQHAS 60
+P D ++ Q F RT P+NT+A+ +R Y++F+ P H LW + I + N H+S
Sbjct 44 IPHDHVEINAQDFMRTLPMNTAAFASMRGVYEFFFVPYHQLWAQFDQFITGM--NDFHSS 101
Query 61 SYDGSVLLGSNMPCVslsqlskllsslkgkkNYFGFDRSDLAYK-------ILQYLRYGN 113
+ + S+ G++ V + + +SL K DL YK +L L YG
Sbjct 102 A-NKSIQGGTSPLQVPYFNVDSVFNSLNTGKESGSGSTDDLQYKFKYGAFRLLDLLGYGR 160
Query 114 VQTSSSTSGKNFGTSIPLSDRSYSQDYVFNHALSIFPLLGYKKFCQDYFRFTQWQDSAPY 173
S FGT+ P + + +N S+F +L Y K QDY+R + +++
Sbjct 161 KFDS-------FGTAYPDNVSGLKNNLDYN--CSVFRILAYNKIYQDYYRNSNYENFDTD 211
Query 174 LWNIDYY-----DAKKSTSILPDTFSTSYLTHNTLIDMEYCNWNKDMFFGVLPDAQYGDA 228
+N D + DAK ++ D F Y N D + N + F + D
Sbjct 212 SFNFDKFKGGLVDAK----VVADLFKLRY--RNAQTDY-FTNLRQSQLFSFT--TAFEDV 262
Query 229 SVVDISFGMSGQTVVASPSDISSRYTISNPSDSSTPNLSGSPLVLDVLALRRGEALQRFR 288
++I + + V S +R +DSS + S V +LR A+ +
Sbjct 263 DNINI----APRDYVKSDGSNFTRVNFGVDTDSSEGDFS-------VSSLRAAFAVDKLL 311
Query 289 EISLCTPANYRSQIKAHFGVDVGSELSGMSTYIGGEASSLDISEVVNTNITESNE----- 343
+++ ++ Q++AH+GV++ G Y+GG S + +S+V T+ T + E
Sbjct 312 SVTMRAGKTFQDQMRAHYGVEIPDSRDGRVNYLGGFDSDMQVSDVTQTSGTTATEYKPEA 371
Query 344 ---ALIAGKGIGTGQFSDKFYAKDWGILMCIYHSVPLLDYVLTSPDPQLFLSENTSFPVP 400
+AGKG G+G+ F AK+ G+LMCIY VP + Y T DP + + + P
Sbjct 372 GYLGRVAGKGTGSGRGRIVFDAKEHGVLMCIYSLVPQIQYDCTRLDPMVDKLDRFDYFTP 431
Query 401 ELDAIGLESIPL-SCYSNSSLEIPITNPNVDAASLTMGYLPRYYAWKTSLDYVLGAFTTT 459
E + +G++ PL S Y +S NP +GY PRY +KT+LD G F +
Sbjct 432 EFENLGMQ--PLNSSYISSFCTTDPKNP-------VLGYQPRYSEYKTALDVNHGQFAQS 482
Query 460 EKEWVAPITASLWSKMLLPVTVDGSGINYNFFKVNPSILDPIFLVNADSTWDTDTFLVNA 519
+ S WS + FK++P L+ IF V+ + T D
Sbjct 483 D-------ALSSWSVSRFRRWTTFPQLEIADFKIDPGCLNSIFPVDYNGTEANDCVYGGC 535
Query 520 AFDIRVARNLDYDGMP 535
F+I ++ DGMP
Sbjct 536 NFNIVKVSDMSVDGMP 551
>gi|517172762|ref|WP_018361580.1| hypothetical protein [Prevotella nanceiensis]
Length=568
Score = 147 bits (370), Expect = 8e-35, Method: Compositional matrix adjust.
Identities = 147/566 (26%), Positives = 237/566 (42%), Gaps = 77/566 (14%)
Query 2 PGDKFSLSEQHFTRTQPVNTSAYTRVREYYDWFWCPLHLLWRNAPEVIAQIQQNVQHASS 61
P D ++ F RT P+N++A+ +R Y++++ P LW + Q + S
Sbjct 46 PHDHVEINASDFMRTLPMNSAAFMSMRGVYEFYFVPYKQLW-------SGFDQFITGMSD 98
Query 62 YDGSVLL---GSNMP-CVslsqlskll-sslkgkkNYFGFDRSDLAYKILQYLRYGNVQT 116
Y S + G P CVS + K+ GFD++ Y+IL L YG
Sbjct 99 YKSSFMYAFKGKTPPSCVSFDVQKLVDWCKTNTAKDIHGFDKNKGVYRILDLLGYGKYAN 158
Query 117 SSSTSGKNFGTSIPLSDRSYSQDYVFNHALSIFPLLGYKKFCQDYFRFTQWQDSAPYLWN 176
S+ N TS + + F L Y+K D++R T +++ +N
Sbjct 159 SAGVPYTN-PTSTTMG------------KCTPFRGLAYQKIYNDFYRNTTYEEYQLESFN 205
Query 177 ID-YYDAKKSTSILPDT-FSTSYLTHNTLIDMEYCNWNKDMFFGVLPDAQYGDASVVDIS 234
+D +Y + K +P+ + + T + Y N KD+ V P + S+ D +
Sbjct 206 VDMFYGSGKVKETIPNEPWDYDWFT------LRYRNAQKDLLTNVRPTPLF---SIDDFN 256
Query 235 --FGMSGQTVV--ASPSDISSRYTISNPSDSSTPNLSGSPL-----VLDVLALRRGEALQ 285
F G +V P+ + + NL + + ++ V +R AL+
Sbjct 257 PQFFTGGSDIVMEKGPNVTGGTHEYRDSVVIVGKNLKENGVDSKRTMISVADIRNAFALE 316
Query 286 RFREISLCTPANYRSQIKAHFGVDVGSELSGMSTYIGGEASSLDISEVVN---TNITESN 342
+ +++ Y+ Q++AHFG+ V G TYIGG S++ + +V T +T +
Sbjct 317 KLASVTMRAGKTYKEQMEAHFGISVEEGRDGRCTYIGGFDSNIQVGDVTQSSGTTVTGTK 376
Query 343 E-------ALIAGKGIGTGQFSDKFYAKDWGILMCIYHSVPLLDYVLTSPDPQLFLSENT 395
+ GK G+G +F AK+ GILMCIY VP + Y DP + E
Sbjct 377 DTSFGGYLGRTTGKATGSGSGHIRFDAKEHGILMCIYSLVPDVQYDSKRVDPFVQKIERG 436
Query 396 SFPVPELDAIGLESIPLSC------YSNSSLEIPITNPNVDAASLTMGYLPRYYAWKTSL 449
F VPE + +G++ PL Y+N++ I N G+ PRY +KT+L
Sbjct 437 DFFVPEFENLGMQ--PLFAKNISYKYNNNTANSRIKNLGA------FGWQPRYSEYKTAL 488
Query 450 DYVLGAFTTTEKEWVAPITASLWSKMLLPVTVDGSGINYNFFKVNPSILDPIFLVNADST 509
D G F E + + M S N + FK+NP LD +F VN + T
Sbjct 489 DINHGQFVHQEPLSYWTVARARGESM--------SNFNISTFKINPKWLDDVFAVNYNGT 540
Query 510 WDTDTFLVNAAFDIRVARNLDYDGMP 535
TD F+I ++ DGMP
Sbjct 541 ELTDQVFGGCYFNIVKVSDMSIDGMP 566
Lambda K H a alpha
0.318 0.134 0.413 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 3854736288630