bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: All non-redundant GenBank CDS translations+PDB+SwissProt+PIR+PRF
excluding environmental samples from WGS projects
49,011,213 sequences; 17,563,301,199 total letters
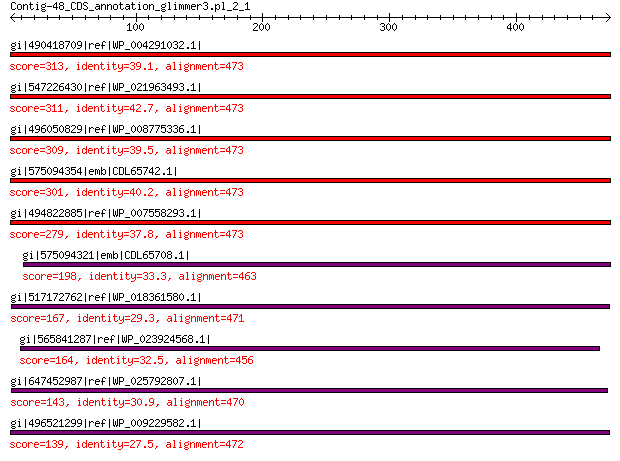
Query= Contig-48_CDS_annotation_glimmer3.pl_2_1
Length=473
Score E
Sequences producing significant alignments: (Bits) Value
gi|490418709|ref|WP_004291032.1| hypothetical protein 313 4e-96
gi|547226430|ref|WP_021963493.1| putative uncharacterized protein 311 9e-96
gi|496050829|ref|WP_008775336.1| hypothetical protein 309 9e-95
gi|575094354|emb|CDL65742.1| unnamed protein product 301 2e-91
gi|494822885|ref|WP_007558293.1| hypothetical protein 279 9e-83
gi|575094321|emb|CDL65708.1| unnamed protein product 198 1e-52
gi|517172762|ref|WP_018361580.1| hypothetical protein 167 6e-42
gi|565841287|ref|WP_023924568.1| hypothetical protein 164 6e-41
gi|647452987|ref|WP_025792807.1| hypothetical protein 143 6e-34
gi|496521299|ref|WP_009229582.1| capsid protein 139 1e-32
>gi|490418709|ref|WP_004291032.1| hypothetical protein [Bacteroides eggerthii]
gi|217986636|gb|EEC52970.1| putative capsid protein (F protein) [Bacteroides eggerthii DSM
20697]
Length=578
Score = 313 bits (801), Expect = 4e-96, Method: Compositional matrix adjust.
Identities = 185/480 (39%), Positives = 259/480 (54%), Gaps = 53/480 (11%)
Query 1 LGFSGADLAYKLLSYLGYGNFIPSPPSNSTRWWSTSLKKEASPTGYTQQYIQNNYVNLFP 60
G++ + + KLL YLGYGN+ S T W+T+ + N N+F
Sbjct 145 FGYNRSKSSVKLLEYLGYGNY----ESFLTDDWNTA------------PLMANLNHNIFG 188
Query 61 LLAYQKIYQDFFRWSQWERANPSSYNVDYYSGVSSSLVTVLPDYTSDYWKSDTMFDLKYC 120
LLAYQKIY DF+R SQWER +PS++NVDY G S +L Y+++++++ FDL+YC
Sbjct 189 LLAYQKIYSDFYRDSQWERVSPSTFNVDYLDGSSMNLDNA---YSTEFYQNYNFFDLRYC 245
Query 121 NWNKDMLMGILPDSQFGECCYKSIFETPGGDLKAGFRTTDGKFISAVTNAPLTTENSSSG 180
NW KD+ G+LP Q+GE SI G L +T G T+ ++SG
Sbjct 246 NWQKDLFHGVLPHQQYGETAVASITPDVTGKLTLSNFSTVG-----------TSPTTASG 294
Query 181 LSTPGVTSGSTVALKSPLISDLSALQSQFSVLALRQAEALQRWKEISQSGDSDYREQIRK 240
+T + + TV S+L LRQAE LQ+WKEI+QSG+ DY++Q+ K
Sbjct 295 TATKNLPAFDTVG--------------DLSILVLRQAEFLQKWKEITQSGNKDYKDQLEK 340
Query 241 HFGVNLPQSLSNLCTYIGGISRNLDISEVVNNNLAAEGDTAVIAGKGVGAGNGSFTYTTD 300
H+GV++ S LCTY+GG+S ++DI+EV+N N+ A IAGKGVG NG + ++
Sbjct 341 HWGVSVGDGFSELCTYLGGVSSSIDINEVINTNITGSA-AADIAGKGVGVANGEINFNSN 399
Query 301 -EHCVVMCIYHAVPLLDYTITGQDGQLLVTDAESLPIPEFDNIGMEVLPMTQIFNSPKAS 359
+ ++MCIYH +PLLDYT D L ++ IPEFD +GM+ +P+ Q+ N P S
Sbjct 400 GRYGLIMCIYHCLPLLDYTTDMLDPAFLKVNSTDYAIPEFDRVGMQSMPLVQLMN-PLRS 458
Query 360 IVNL--FNAGYNPRYFNWKTKLDVVNGAFTTTLKSWVSPVTESLLSGWFGFGYSQDDVNK 417
N GY PRY ++KT +D G F TL SWV + +
Sbjct 459 FANASGLVLGYVPRYIDYKTSVDQSVGGFKRTLNSWVISYGNISVLKQVTLPNDAPPIEP 518
Query 418 DTKV----VLNYKFFKVNPSVLDPIFGVNADSTWDTDQLLVNSYIGCYVARNLSRDGVPY 473
V +N+ FFKVNP LDPIF V A +TDQ L +S+ RNL DG+PY
Sbjct 519 SEPVPSVAPMNFTFFKVNPDCLDPIFAVQAGDDTNTDQFLCSSFFDIKAVRNLDTDGLPY 578
>gi|547226430|ref|WP_021963493.1| putative uncharacterized protein [Prevotella sp. CAG:1185]
gi|524103382|emb|CCY83994.1| putative uncharacterized protein [Prevotella sp. CAG:1185]
Length=573
Score = 311 bits (798), Expect = 9e-96, Method: Compositional matrix adjust.
Identities = 202/484 (42%), Positives = 267/484 (55%), Gaps = 64/484 (13%)
Query 1 LGFSGADLAYKLLSYLGYGNFIPSPPSNSTRWWSTSLKKEASPTGYTQQYIQNNYVNLFP 60
GFS +L+ KLL+YL YG S + S S SP FP
Sbjct 143 FGFSRVELSVKLLNYLNYGF---GKDYESVKVPSDSDDIVLSP---------------FP 184
Query 61 LLAYQKIYQDFFRWSQWERANPSSYNVDYYSGVSSSLVTVLPDYTSDYWKSDTMFDLKYC 120
LLAYQKI +D+FR QW+ A P YN+DY G SS + +T+D +K+ TMFDL YC
Sbjct 185 LLAYQKICEDYFRDDQWQSAAPYRYNLDYLYGKSSGFHIPMSSFTNDAFKNPTMFDLNYC 244
Query 121 NWNKDMLMGILPDSQFGECCYKS-IFETPGGDLKAGFRTTDGKFISAVTNAPLTTENSSS 179
N+ KD G+LP +Q+G+ S IF GDL G ++ F SA T + S
Sbjct 245 NFQKDYFTGMLPRAQYGDVSVASPIF----GDLDIG-DSSSLTFASAPQQGANTIQ---S 296
Query 180 GLSTPGVTSGSTVALKSPLISDLSALQSQFSVLALRQAEALQRWKEISQSGDSDYREQIR 239
G+ S +T L SVLALRQAE LQ+W+EI+QSG DY+ Q++
Sbjct 297 GVLVVNNNSNTTAGL---------------SVLALRQAECLQKWREIAQSGKMDYQTQMQ 341
Query 240 KHFGVNLPQSLSNLCTYIGGISRNLDISEVVNNNLAAEGDTAVIAGKGVGAGNGS-FTYT 298
KHF V+ +LS C Y+GG + NLDISEVVN NL + + A I GKG G NG+ +
Sbjct 342 KHFNVSPSATLSGHCKYLGGWTSNLDISEVVNTNLTGD-NQADIQGKGTGTLNGNKVDFE 400
Query 299 TDEHCVVMCIYHAVPLLDYTITGQDGQLLVTDAESLPIPEFDNIGMEVL-PMTQIFN--- 354
+ EH ++MCIYH +PLLD++I Q T IPEFD++GM+ L P IF
Sbjct 401 SSEHGIIMCIYHCLPLLDWSINRIARQNFKTTFTDYAIPEFDSVGMQQLYPSEMIFGLED 460
Query 355 --SPKASIVNLFNAGYNPRYFNWKTKLDVVNGAFTTTLKSWVSPVTESLLSGWFGFGYSQ 412
S +SI N GY PRY + KT +D ++G+F TL SWVSP+T+S +S +
Sbjct 461 LPSDPSSI----NMGYVPRYADLKTSIDEIHGSFIDTLVSWVSPLTDSYISAY------- 509
Query 413 DDVNKD---TKVVLNYKFFKVNPSVLDPIFGVNADSTWDTDQLLVNSYIGCYVARNLSRD 469
KD + + + Y FFKVNP ++D IFGV ADST +TDQLL+NSY RN +
Sbjct 510 RQACKDAGFSDITMTYNFFKVNPHIVDNIFGVKADSTINTDQLLINSYFDIKAVRNFDYN 569
Query 470 GVPY 473
G+PY
Sbjct 570 GLPY 573
>gi|496050829|ref|WP_008775336.1| hypothetical protein [Bacteroides sp. 2_2_4]
gi|229448893|gb|EEO54684.1| putative capsid protein (F protein) [Bacteroides sp. 2_2_4]
Length=580
Score = 309 bits (792), Expect = 9e-95, Method: Compositional matrix adjust.
Identities = 187/477 (39%), Positives = 265/477 (56%), Gaps = 46/477 (10%)
Query 1 LGFSGADLAYKLLSYLGYGNFIPSPPSNSTRWWSTSLKKEASPTGYTQQYIQNNYVNLFP 60
G+S + KLL YLGYGNF S + W + L N +N++
Sbjct 146 FGYSRSLGTAKLLEYLGYGNFYTYATSKNNTWTKSPLSS-------------NLQLNIYG 192
Query 61 LLAYQKIYQDFFRWSQWERANPSSYNVDYYSGVSSSLVTVLPDYTSD-YWKSDTMFDLKY 119
+LAYQKIY D R SQWE+ +PS +NVDY SG S +T+ T + MFDL+Y
Sbjct 193 VLAYQKIYADHIRDSQWEKVSPSCFNVDYLSGTVDSAMTIDSMITGQGFAPFYNMFDLRY 252
Query 120 CNWNKDMLMGILPDSQFGECCYKSIFETPGGDLKAGFRTTDGKFISAVTNAPLTTENSSS 179
CNW KD+ G+LP Q+G+ ++ + + +T DG V +P SS+
Sbjct 253 CNWQKDLFHGVLPRQQYGDTAAVNVNLSNVLSAQYMVQTPDG---DPVGGSPF----SST 305
Query 180 GLSTPGVTSGSTVALKSPLISDLSALQSQFSVLALRQAEALQRWKEISQSGDSDYREQIR 239
G++ V T F+VLALRQAE LQ+WKEI+QSG+ DY++QI
Sbjct 306 GVNLQTVNGSGT-----------------FTVLALRQAEFLQKWKEITQSGNKDYKDQIE 348
Query 240 KHFGVNLPQSLSNLCTYIGGISRNLDISEVVNNNLAAEGDTAVIAGKGVGAGNGSFTYTT 299
KH+ V++ ++ S + Y+GG + +LDI+EVVNNN+ + A IAGKGV GNG ++
Sbjct 349 KHWNVSVGEAYSEMSLYLGGTTASLDINEVVNNNITGS-NAADIAGKGVVVGNGRISFDA 407
Query 300 DE-HCVVMCIYHAVPLLDYTITGQDGQLLVTDAESLPIPEFDNIGMEVLPMTQIFNSPKA 358
E + ++MCIYH++PLLDYT + ++ IPEFD +GME +P+ + N P
Sbjct 408 GERYGLIMCIYHSLPLLDYTTDLVNPAFTKINSTDFAIPEFDRVGMESVPLVSLMN-PLQ 466
Query 359 SIVNLFNA--GYNPRYFNWKTKLDVVNGAFTTTLKSWVSPVTESLLSGWFGFGYSQDDVN 416
S N+ ++ GY PRY ++KT +D GAF TTLKSWV + + QDD N
Sbjct 467 SSYNVGSSILGYAPRYISYKTDVDSSVGAFKTTLKSWVMSYDNQSVINQLNY---QDDPN 523
Query 417 KDTKVVLNYKFFKVNPSVLDPIFGVNADSTWDTDQLLVNSYIGCYVARNLSRDGVPY 473
++NY FKVNP+ +DP+F V A ++ DTDQ L +S+ V RNL DG+PY
Sbjct 524 NSPGTLVNYTNFKVNPNCVDPLFAVAASNSIDTDQFLCSSFFDVKVVRNLDTDGLPY 580
>gi|575094354|emb|CDL65742.1| unnamed protein product [uncultured bacterium]
Length=615
Score = 301 bits (771), Expect = 2e-91, Method: Compositional matrix adjust.
Identities = 190/506 (38%), Positives = 272/506 (54%), Gaps = 59/506 (12%)
Query 1 LGFSGADLAYKLLSYLGYGNFIPSPPSNSTRWWSTSLKKEASPTGYTQQYIQNNYVNLFP 60
GF+ + L KLL YLGYG++ + + T WS A P Y + ++ FP
Sbjct 136 FGFNRSTLTCKLLQYLGYGDY--NSFDSETNTWS------AKPLLYNLE------LSPFP 181
Query 61 LLAYQKIYQDFFRWSQWERANPSSYNVDYYSGVSSSLVTVLPDYTSDYWKSDTMFDLKYC 120
LLAYQKIY DF+R++QWE+ NPS++N+DY G +S L L SD + FD++YC
Sbjct 182 LLAYQKIYSDFYRYTQWEKTNPSTFNLDYIKG-TSDLQMDLTGLPSD---DNNFFDIRYC 237
Query 121 NWNKDMLMGILPDSQFGECCYKSIFETPGGDLKAGFRTTDGKFISAVTNAPLT------- 173
N+ KDM G+LP +Q+G I G L G T P T
Sbjct 238 NYQKDMFHGVLPVAQYGSASVVPI----NGQLNVISNGDSGPIFKTSTPDPGTPGTSYVT 293
Query 174 ------TENSSSGLSTPGVTSG-----------STVALKSPLISDLSALQSQ-----FSV 211
+N S G+S + G S + +S L + + + +
Sbjct 294 VGGNIGVDNRSFGVSGSTLNVGKSADPSGYGFPSNASTRSLLWENPNLIIENNQGFYVPI 353
Query 212 LALRQAEALQRWKEISQSGDSDYREQIRKHFGVNLPQSLSNLCTYIGGISRNLDISEVVN 271
LALRQAE LQ+WKE+S SG+ DY+ QI KH+G+ + LS+ Y+GG + +LDI+EV+N
Sbjct 354 LALRQAEFLQKWKEVSVSGEEDYKSQIEKHWGIKVSDFLSHQARYLGGCATSLDINEVIN 413
Query 272 NNLAAEGDTAVIAGKGVGAGNGSFTYTTD-EHCVVMCIYHAVPLLDYTITGQDGQLLVTD 330
NN+ + + A IAGKG GNGS + + E+ ++MCIYH +P++DY +G D + D
Sbjct 414 NNITGD-NAADIAGKGTFTGNGSIRFESKGEYGIIMCIYHVLPIVDYVGSGVDHSCTLVD 472
Query 331 AESLPIPEFDNIGMEVLPMTQIFNSPKASIVNLFNA--GYNPRYFNWKTKLDVVNGAFTT 388
A S PIPE D IGME +P+ + N K S + GY PRY +WKT +D G F
Sbjct 473 ATSFPIPELDQIGMESVPLVRAMNPVKESDTPSADTFLGYAPRYIDWKTSVDRSVGDFAD 532
Query 389 TLKSWVSPVTESLLSGWFGFGY-SQDDVNKDTKVVLNYKFFKVNPSVLDPIFGVNADSTW 447
+L++W PV + L+ + S +V D+ + FFKVNPS++DP+F V ADST
Sbjct 533 SLRTWCLPVGDKELTSANSLNFPSNPNVEPDS---IAAGFFKVNPSIVDPLFAVVADSTV 589
Query 448 DTDQLLVNSYIGCYVARNLSRDGVPY 473
TD+ L +S+ V RNL +G+PY
Sbjct 590 KTDEFLCSSFFDVKVVRNLDVNGLPY 615
>gi|494822885|ref|WP_007558293.1| hypothetical protein [Bacteroides plebeius]
gi|198272099|gb|EDY96368.1| putative capsid protein (F protein) [Bacteroides plebeius DSM
17135]
Length=613
Score = 279 bits (713), Expect = 9e-83, Method: Compositional matrix adjust.
Identities = 179/487 (37%), Positives = 265/487 (54%), Gaps = 32/487 (7%)
Query 1 LGFSGADLAYKLLSYLGYGNFIPSPPSNSTRWWSTSLKKEASPTGYTQQYIQNNYVNLFP 60
G+ A L +L YLGYG+F P + T T+ + N + FP
Sbjct 145 FGYYRAWLVCIILEYLGYGDFYP---------YIVEAAGGEGATWATRPMLNNLKFSPFP 195
Query 61 LLAYQKIYQDFFRWSQWERANPSSYNVDYYSGVSSSLVTVLPDYTSDYWK-SDTMFDLKY 119
L AYQKIY DF R++QWER+NPS++N+DY SG + SL D+T + +K S +FD++Y
Sbjct 196 LFAYQKIYADFNRYTQWERSNPSTFNIDYISGSADSLQL---DFTVEGFKDSFNLFDMRY 252
Query 120 CNWNKDMLMGILPDSQFGECCYKSI---FETPGGDLKAGFRTTDGKFISAVTNAPLTTEN 176
NW +D+L G +P +Q+GE + + G F T G+ A N +T +
Sbjct 253 SNWQRDLLHGTIPQAQYGEASAVPVSGSMQVVEGPTPPAFTT--GQDGVAFLNGNVTIQG 310
Query 177 SSSGLSTPGVTSGSTVALKSPLISDLSAL-QSQF--SVLALRQAEALQRWKEISQSGDSD 233
SS L S + + S L S F S+LALR+AEA Q+WKE++ + + D
Sbjct 311 SSGYLQAQTSVGESRILRFNNTNSGLIVEGDSSFGVSILALRRAEAAQKWKEVALASEED 370
Query 234 YREQIRKHFGVNLPQSLSNLCTYIGGISRNLDISEVVNNNLAAEGDTAVIAGKGVGAGNG 293
Y QI H+G ++ ++ S++C ++G I+ +L I+EVVNNN+ E + A IAGKG +GNG
Sbjct 371 YPSQIEAHWGQSVNKAYSDMCQWLGSINIDLSINEVVNNNITGE-NAADIAGKGTMSGNG 429
Query 294 SFTYTT-DEHCVVMCIYHAVPLLDYTITGQDGQLLVTDAESLPIPEFDNIGMEVLPMTQI 352
S + ++ +VMC++H +P LDY + +T+ PIPEFD IGME +P+ +
Sbjct 430 SINFNVGGQYGIVMCVFHVLPQLDYITSAPHFGTTLTNVLDFPIPEFDKIGMEQVPVIRG 489
Query 353 FNSPKAS------IVNLFNAGYNPRYFNWKTKLDVVNGAFTTTLKSWVSPVTESLLSGWF 406
N K NL+ GY P+Y+NWKT LD G F +LK+W+ P + L
Sbjct 490 LNPVKPKDGDFKVSPNLY-FGYAPQYYNWKTTLDKSMGEFRRSLKTWIIPFDDEALLAAD 548
Query 407 GFGYSQDDVNKDTKVVLNYKFFKVNPSVLDPIFGVNADSTWDTDQLLVNSYIGCYVARNL 466
+ D+ N + V FFKV+PSVLD +F V A+S +TDQ L ++ V R+L
Sbjct 549 SVDFP-DNPNVEADSV-KAGFFKVSPSVLDNLFAVKANSDLNTDQFLCSTLFDVNVVRSL 606
Query 467 SRDGVPY 473
+G+PY
Sbjct 607 DPNGLPY 613
>gi|575094321|emb|CDL65708.1| unnamed protein product [uncultured bacterium]
Length=642
Score = 198 bits (503), Expect = 1e-52, Method: Compositional matrix adjust.
Identities = 154/502 (31%), Positives = 234/502 (47%), Gaps = 66/502 (13%)
Query 11 KLLSYLGYGNFIPSPPSNSTRWWSTSLKKEASPTGYTQQYIQNN-YVNLFPLLAYQKIYQ 69
KLL LGYGNF P + + K S + N+ Y+++F LLAY KI
Sbjct 166 KLLQLLGYGNF----PEQFANFKVNNDKHNQSGQNFKDVTYNNSPYLSIFRLLAYHKICN 221
Query 70 DFFRWSQWERANPSSYNVDYYSGVSSSLVTV------LPDYTSDYWKSD--TMFDLKYCN 121
D + + QW+ N S NVDY + SSSL+++ +PD D K++ + D+++ N
Sbjct 222 DHYLYRQWQPYNASLCNVDYLTPNSSSLLSIDDALLSIPD---DSIKAEKLNLLDMRFSN 278
Query 122 WNKDMLMGILPDSQFG----------ECCYKSIFETPGGDLKAGFRTTDGKF-----ISA 166
D G+LP SQFG ++ +RTT G++ +++
Sbjct 279 LPLDYFTGVLPTSQFGSESVVNLNLGNASGSAVLNGTTSKDSGRWRTTTGEWEMEQRVAS 338
Query 167 VTNAPLTTENSSSGLSTPGVTSGSTVALKSPLISDLSALQSQFSVLALRQAEALQRWKEI 226
N L +NS+ + T VA+ + L +LS ++ALR A A Q++KEI
Sbjct 339 SANGNLKLDNSNGTFISHDHTFSGNVAINTSLSGNLS-------IIALRNALAAQKYKEI 391
Query 227 SQSGDSDYREQIRKHFGVNLPQSLSNLCTYIGGISRNLDISEVVNNNLAAEGDTAVIAGK 286
+ D D++ Q+ HFG+ P + +IGG S ++I+E +N NL+ + + A
Sbjct 392 QLANDVDFQSQVEAHFGIK-PDEKNENSLFIGGSSSMININEQINQNLSGD-NKATYGAA 449
Query 287 GVGAGNGSFTYTTDEHCVVMCIYHAVPLLDYTITGQDGQLLVTDAESLPIPEFDNIGMEV 346
G G+ S +T + VV+ IY P+LD+ G D L TDA IPE D+IGM+
Sbjct 450 PQGNGSASIKFTAKTYGVVIGIYRCTPVLDFAHLGIDRTLFKTDASDFVIPEMDSIGMQQ 509
Query 347 LPMTQIFNSPKASIVNLFNA---------------GYNPRYFNWKTKLDVVNGAFTTTLK 391
++ + A + F A GY PRY +KT D NGAF +LK
Sbjct 510 TFRCEV--AAPAPYNDEFKAFRVGDGSSPDMSETYGYAPRYSEFKTSYDRYNGAFCHSLK 567
Query 392 SWVSPVTESLLSGWFGFGYSQDDVNKDTKVVLNYKFFKVNPSVLDPIFGVNADSTWDTDQ 451
SWV+ + F Q++V + F P ++ +F V++ + D DQ
Sbjct 568 SWVTGIN---------FDAIQNNVWNTWAGINAPNMFACRPDIVKNLFLVSSTNNSDDDQ 618
Query 452 LLVNSYIGCYVARNLSRDGVPY 473
L V CY RNLSR G+PY
Sbjct 619 LYVGMVNMCYATRNLSRYGLPY 640
>gi|517172762|ref|WP_018361580.1| hypothetical protein [Prevotella nanceiensis]
Length=568
Score = 167 bits (422), Expect = 6e-42, Method: Compositional matrix adjust.
Identities = 138/493 (28%), Positives = 211/493 (43%), Gaps = 85/493 (17%)
Query 2 GFSGADLAYKLLSYLGYGNFIPSPPSNSTRWWSTSLKKEASPTGYTQQYIQNNYVNLFPL 61
GF Y++L LGYG + S T ST++ K +P F
Sbjct 137 GFDKNKGVYRILDLLGYGKYANSAGVPYTNPTSTTMGK-CTP---------------FRG 180
Query 62 LAYQKIYQDFFRWSQWERANPSSYNVDYYSGVSSSLVTVLPDYTSDY-WKSDTMFDLKYC 120
LAYQKIY DF+R + +E S+NVD + G S + +P+ DY W F L+Y
Sbjct 181 LAYQKIYNDFYRNTTYEEYQLESFNVDMFYG-SGKVKETIPNEPWDYDW-----FTLRYR 234
Query 121 NWNKDMLMGILPDSQFGECCYKSIFETPGGDLKAGFRTTDGKFISAVTNAPLTTENSSSG 180
N KD+L + P F + F T G D + E
Sbjct 235 NAQKDLLTNVRPTPLFSIDDFNPQFFTGGSD--------------------IVMEKG--- 271
Query 181 LSTPGVTSGSTVALKSPLI-------SDLSALQSQFSVLALRQAEALQRWKEISQSGDSD 233
P VT G+ S +I + + + ++ SV +R A AL++ ++
Sbjct 272 ---PNVTGGTHEYRDSVVIVGKNLKENGVDSKRTMISVADIRNAFALEKLASVTMRAGKT 328
Query 234 YREQIRKHFGVNLPQSLSNLCTYIGGISRNLDISEVVNNN---LAAEGDTAV------IA 284
Y+EQ+ HFG+++ + CTYIGG N+ + +V ++ + DT+
Sbjct 329 YKEQMEAHFGISVEEGRDGRCTYIGGFDSNIQVGDVTQSSGTTVTGTKDTSFGGYLGRTT 388
Query 285 GKGVGAGNGSFTYTTDEHCVVMCIYHAVPLLDYTITGQDGQLLVTDAESLPIPEFDNIGM 344
GK G+G+G + EH ++MCIY VP + Y D + + +PEF+N+GM
Sbjct 389 GKATGSGSGHIRFDAKEHGILMCIYSLVPDVQYDSKRVDPFVQKIERGDFFVPEFENLGM 448
Query 345 EVLPMTQIF-----NSPKASIVNLFNAGYNPRYFNWKTKLDVVNGAFTTTLKSWVSPVTE 399
+ L I N+ + I NL G+ PRY +KT LD+ +G F V +
Sbjct 449 QPLFAKNISYKYNNNTANSRIKNLGAFGWQPRYSEYKTALDINHGQF----------VHQ 498
Query 400 SLLSGWFGFGYSQDDVNKDTKVVLNYKFFKVNPSVLDPIFGVNADSTWDTDQLLVNSYIG 459
LS W + ++ N FK+NP LD +F VN + T TDQ+ Y
Sbjct 499 EPLSYW-----TVARARGESMSNFNISTFKINPKWLDDVFAVNYNGTELTDQVFGGCYFN 553
Query 460 CYVARNLSRDGVP 472
++S DG+P
Sbjct 554 IVKVSDMSIDGMP 566
>gi|565841287|ref|WP_023924568.1| hypothetical protein [Prevotella nigrescens]
gi|564729907|gb|ETD29851.1| hypothetical protein HMPREF1173_00033 [Prevotella nigrescens
CC14M]
Length=656
Score = 164 bits (416), Expect = 6e-41, Method: Compositional matrix adjust.
Identities = 148/477 (31%), Positives = 216/477 (45%), Gaps = 70/477 (15%)
Query 9 AYKLLSYLGYG----NFIPSPPSNSTRWWSTSLKKEASPTGYTQQYIQNNYVNLFPLLAY 64
A++LL +LGYG FI ++ +K + Y I+ N+F LLAY
Sbjct 219 AFRLLHFLGYGVDNNGFIVDFNASYAAGTGEIVKNVLAKKTYKLPDIK---ANVFRLLAY 275
Query 65 QKIYQDFFRWSQWERANPSSYNVDYYSGVSSSLVTVLPDYTSDYWKSDTMFDLKYCNWNK 124
Q+IY DF+R WE A P +NVD+ +S ++ Y M L+Y +W+K
Sbjct 276 QRIYNDFYRNDLWEAAQPDVFNVDWCCNNNSLDISDELVY--------KMCQLRYRHWSK 327
Query 125 DMLMGILPDSQFGECCYKSIFETPGG-DLKAGFRTTDGKFISAVTNAPLTTENSSSGLST 183
D + P + + K IFE P + GF TT+ K V N N S L
Sbjct 328 DWVTSAYPTASYD----KGIFELPDYINGNTGFATTEVK--RDVVN------NRGSQLEI 375
Query 184 PGVTSGSTVALKSPLISDLSALQSQFSVLALRQAEALQRWKEISQSGDS-DYREQIRKHF 242
+ +GS L S IS +S +R AL++ E +++ + DY QI HF
Sbjct 376 KSMDAGS---LGSNNISYISPND-------IRAMFALEKMLERTRAANGLDYSNQIAAHF 425
Query 243 GVNLPQSLSNLCTYIGGISRNLDISEVVNNNLAAEGDTAV-------IAGKGVGAGN-GS 294
G +P+S N ++IGG + ISEVV + + TA + GKG+GA N G
Sbjct 426 GFKVPESRKNCASFIGGFDNQISISEVVTTSNGSVDGTASTGSVVGQVFGKGIGAMNSGH 485
Query 295 FTYTTDEHCVVMCIYHAVPLLDYTITGQDGQLLVTDAESLPIPEFDNIGMEVLPMTQI-- 352
+Y EH ++MCIY P +DY D E PEF+N+GM+ + + +
Sbjct 486 ISYDVKEHGLIMCIYSIAPQVDYDARELDPFNRKFSREDYFQPEFENLGMQPVIQSDLCL 545
Query 353 -FNSPKASIVNLFN--AGYNPRYFNWKTKLDVVNGAFTT--TLKSWVSPVTESLLSGWFG 407
NS K+ + N GY+ RY +KT D++ G F + +L +W +P F
Sbjct 546 CINSAKSDSSDQHNNVLGYSARYLEYKTARDIIFGEFMSGGSLSAWATPKNNYT----FE 601
Query 408 FGYSQDDVNKDTKVVLNYKFFKVNPSVLDPIFGVNADSTWDTDQLLVNSYIGCYVAR 464
FG L+ V+P VL+PIF V + + TDQ LVNSY R
Sbjct 602 FG------------KLSLPDLLVDPKVLEPIFAVKYNGSMSTDQFLVNSYFDVKAIR 646
>gi|647452987|ref|WP_025792807.1| hypothetical protein [Prevotella histicola]
Length=584
Score = 143 bits (361), Expect = 6e-34, Method: Compositional matrix adjust.
Identities = 145/511 (28%), Positives = 229/511 (45%), Gaps = 104/511 (20%)
Query 2 GFSGADLAYKLLSYLGYG-----------NFIPSPPSNSTRWWSTSLKKEASPTGYTQQY 50
GF+ ++ A KLL+ L YG N I ST + + KE S
Sbjct 134 GFNYSEGAAKLLNMLNYGVTNKGKFMNLENLI-----TSTSYLPSKDDKEPSS------- 181
Query 51 IQNNYVNLFPLLAYQKIYQDFFRWSQWERANPSSYNVDYYSGVSSSLVTVLPDYTSDYWK 110
I V+ F LLAYQKI+ DF+R W ++ S+NVD Y+ S+ +T+ PD + +
Sbjct 182 IYACKVSPFRLLAYQKIFNDFYRNQDWTPSDVRSFNVDDYADDSN--LTIEPDVALKFCQ 239
Query 111 SDTMFDLKYCNWNKDMLMGILPDSQFGECCYKSIFETPGGDLKAGFRTTDGKFISAVTNA 170
++Y + KD L + P + + IF P +++ N
Sbjct 240 ------MRYRPYAKDWLTSMKPTPNYSD----GIFNLP-------------EYVRGNGNV 276
Query 171 PLTTENSSSGLSTPGVTSGSTVALKSPLISDLSALQSQFSVLALRQAEALQRWKEISQSG 230
LT S S V+L S +S S FSV LR A AL + E ++
Sbjct 277 ILTNNKSGS------------VSLDSGTVS-----PSSFSVNDLRAAFALDKMLEATRRA 319
Query 231 DS-DYREQIRKHFGVNLPQSLSNLCTYIGGISRNLDISEVV--NNNLAAEGDTAVI---A 284
+ DY QI HFG +P+S +N ++GG ++ +SEVV N N A++G A I
Sbjct 320 NGLDYASQIEAHFGFKVPESRANDARFLGGFDNSIVVSEVVSTNGNAASDGSHASIGDLG 379
Query 285 GKGVGA-GNGSFTYTTDEHCVVMCIYHAVPLLDYTITGQDGQLLVTDAESLPIPEFDNIG 343
GKG+G+ +G+ + + EH ++MCIY P +Y + D E PEF ++G
Sbjct 380 GKGIGSMSSGTIEFDSTEHGIIMCIYSVAPQSEYNASYLDPFNRKLTREQFYQPEFADLG 439
Query 344 MEVLPMTQI------FNSPKA--SIVNLFN--AGYNPRYFNWKTKLDVVNGAFTT--TLK 391
+ L + + N +A S + L N GY RY +KT D+V G F + +L
Sbjct 440 YQALIGSDLICSTLGMNEKQAGFSDIELNNNLLGYQVRYNEYKTARDLVFGDFESGKSLS 499
Query 392 SWVSPVTESLLSGWFGFGYSQDDVNKDTKVVLNYKF-----------FKVNPSVLDPIFG 440
W +P + FG+G ++ + + K +Y+ F +NP++++PIF
Sbjct 500 YWCTPRFD------FGYGDTEKKIAPENKGGADYRKKGNRSHWSSRNFYINPNLVNPIFL 553
Query 441 VNADSTWDTDQLLVNSYIGCYVARNLSRDGV 471
+A D +VNS++ R +S G+
Sbjct 554 TSA---VQADHFIVNSFLDVKAVRPMSVTGL 581
>gi|496521299|ref|WP_009229582.1| capsid protein [Prevotella sp. oral taxon 317]
gi|288330570|gb|EFC69154.1| putative capsid protein (F protein) [Prevotella sp. oral taxon
317 str. F0108]
Length=541
Score = 139 bits (350), Expect = 1e-32, Method: Compositional matrix adjust.
Identities = 130/486 (27%), Positives = 203/486 (42%), Gaps = 97/486 (20%)
Query 1 LGFSGADLAYKLLSYLGYGNFIPSPPSNSTRWWSTSLKKEASPTGYTQQYIQNNYVNLFP 60
G+ ++ + +L+ LGYG I S K P YT VNLF
Sbjct 137 FGYPHSNNSCRLMDLLGYGKPITS-------------SKTPVPLLYTGN------VNLFR 177
Query 61 LLAYQKIYQDFFRWSQWERANPSSYNVDYYSGVSSSLVTVLPDYTSDYWKSDTMFDLKYC 120
LLAY KIY D++R + +E + S+N+D+ G T +P T+D +K +L Y
Sbjct 178 LLAYNKIYSDYYRNTTYEGVDVYSFNIDHKKG------TFVP--TADEFKK--YLNLHYR 227
Query 121 NWNKDMLMGILPDSQF--GECCYKSIFETPGGDLKAGFRTTDGKFISAVTNAPLTTENSS 178
N D + P F G + S+ + L+ S
Sbjct 228 NAPLDFYTNLRPTPLFTIGSDSFSSVLQ-------------------------LSDPTGS 262
Query 179 SGLSTPGVTSGSTVALKSPLISDLSALQSQFSVLALRQAEALQRWKEISQSGDSDYREQI 238
+G S G + + + + SP + ++SA++S F AL + IS Y EQI
Sbjct 263 AGFSADG--NSAKLNMASPDVLNVSAIRSAF---------ALDKLLSISMRAGKTYAEQI 311
Query 239 RKHFGVNLPQSLSNLCTYIGGISRNLDISEV------VNNNLAAEGDTAV------IAGK 286
HFGV + + Y+GG N+ + +V N N++ G+ + I GK
Sbjct 312 EAHFGVTVSEGRDGQVYYLGGFDSNVQVGDVTQTSGTTNPNVSEVGNAKLAGYLGKITGK 371
Query 287 GVGAGNGSFTYTTDEHCVVMCIYHAVPLLDYTITGQDGQLLVTDAESLPIPEFDNIGMEV 346
G G+G G + E V+MCIY VP + Y D + IPEF+N+GM+
Sbjct 372 GTGSGYGEIQFDAKEPGVLMCIYSVVPAMQYDCMRLDPFVAKQTRGDYFIPEFENLGMQ- 430
Query 347 LPMTQIFNSPKASIVNLFNAGYNPRYFNWKTKLDVVNGAFTTTLKSWVSPVTESLLSGWF 406
P+ F S + N + G+ PRY +KT D+ +G F P++ ++
Sbjct 431 -PIVPAFVSLNRAKDNSY--GWQPRYSEYKTAFDINHGQFANG-----EPLSYWSIARAR 482
Query 407 GFGYSQDDVNKDTKVVLNYKFFKVNPSVLDPIFGVNADSTWDTDQLLVNSYIGCYVARNL 466
G DT N K+NP LD +F VN + T TD + ++ ++
Sbjct 483 G---------SDTLNTFNVAALKINPHWLDSVFAVNYNGTEVTDCMFGYAHFNIEKVSDM 533
Query 467 SRDGVP 472
+ DG+P
Sbjct 534 TEDGMP 539
Lambda K H a alpha
0.317 0.134 0.410 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 3246464580183