bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: All non-redundant GenBank CDS translations+PDB+SwissProt+PIR+PRF
excluding environmental samples from WGS projects
49,011,213 sequences; 17,563,301,199 total letters
Query= Contig-4_CDS_annotation_glimmer3.pl_2_2
Length=598
Score E
Sequences producing significant alignments: (Bits) Value
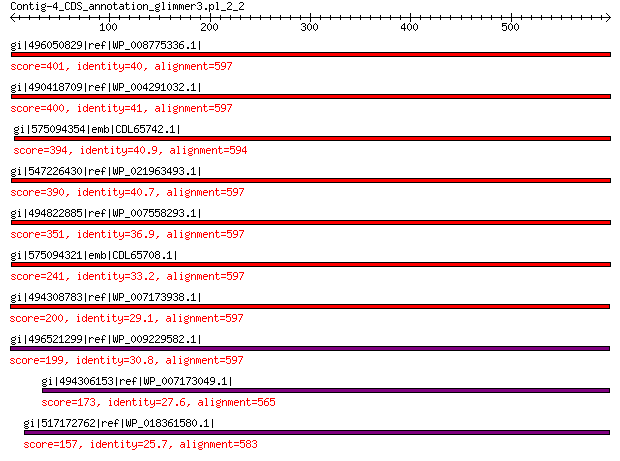
gi|496050829|ref|WP_008775336.1| hypothetical protein 401 2e-128
gi|490418709|ref|WP_004291032.1| hypothetical protein 400 5e-128
gi|575094354|emb|CDL65742.1| unnamed protein product 394 3e-125
gi|547226430|ref|WP_021963493.1| putative uncharacterized protein 390 2e-124
gi|494822885|ref|WP_007558293.1| hypothetical protein 351 1e-108
gi|575094321|emb|CDL65708.1| unnamed protein product 241 7e-67
gi|494308783|ref|WP_007173938.1| hypothetical protein 200 8e-53
gi|496521299|ref|WP_009229582.1| capsid protein 199 1e-52
gi|494306153|ref|WP_007173049.1| hypothetical protein 173 1e-43
gi|517172762|ref|WP_018361580.1| hypothetical protein 157 4e-38
>gi|496050829|ref|WP_008775336.1| hypothetical protein [Bacteroides sp. 2_2_4]
gi|229448893|gb|EEO54684.1| putative capsid protein (F protein) [Bacteroides sp. 2_2_4]
Length=580
Score = 401 bits (1030), Expect = 2e-128, Method: Compositional matrix adjust.
Identities = 239/620 (39%), Positives = 352/620 (57%), Gaps = 65/620 (10%)
Query 2 SLFSLKDIRNHPRRSAFDLSSKVAFSAKSGELLPIKWYFTMPGDKFTLKRQHFTRTQPVN 61
++ SLK +RN R+ FDLSSK F+AK GELLP+K + +PGDK+++ + FTRTQP+N
Sbjct 3 NIMSLKSLRNKTSRNGFDLSSKRNFTAKPGELLPVKCWEVLPGDKWSIDLKSFTRTQPLN 62
Query 62 TSAYTRIREYYDWFWVPLHLLWRHAPEVISQMQSNVQHAGS--QTSSLTLGNYLPTISSS 119
T+A+ R+REYYD+++VP +LLW A V++QM N QHA S +++ L +P ++
Sbjct 63 TAAFARMREYYDFYFVPYNLLWNKANTVLTQMYDNPQHATSYIPSANQALAGVMPNVTCK 122
Query 120 QLSA--------VCSRLFGKKNYFGYDRSDLSYKLMQYLRVGNSGQVSVNFGTSLPASDT 171
++ V + +KNYFGY RS + KL++YL GN F T + +
Sbjct 123 GIADYLNLVAPDVTTTNSYEKNYFGYSRSLGTAKLLEYLGYGN-------FYTYATSKNN 175
Query 172 SYTQA-YRFNLDLSLFPFLAYKKFCQDYFRYSQWQDSSPYLWNIDYFTGvsshlfsslpv 230
++T++ NL L+++ LAY+K D+ R SQW+ SP +N+DY +G + +
Sbjct 176 TWTKSPLSSNLQLNIYGVLAYQKIYADHIRDSQWEKVSPSCFNVDYLSGTVDSAMTIDSM 235
Query 231 ssD----PYWNNNTLFDLEYCNWNKDMFMGVFPDTQFGDVATIGITSDSPESSLQLKAWa 286
+ P++N +FDL YCNW KD+F GV P Q+GD A + + + S+
Sbjct 236 ITGQGFAPFYN---MFDLRYCNWQKDLFHGVLPRQQYGDTAAVNVNLSNVLSA------- 285
Query 287 sgspsskapvvvgaaasspNFTIRAESGN---MNPANILGVDTSSLSLAGSFDVLALRRG 343
+ ++ G+ +P + GV+ +++ +G+F VLALR+
Sbjct 286 -------------------QYMVQTPDGDPVGGSPFSSTGVNLQTVNGSGTFTVLALRQA 326
Query 344 EALQRWKEISLNVPQNYRAQIKAHFGVDVGENMSGMSTYVGGDSSSLDISEVVNTNLQSG 403
E LQ+WKEI+ + ++Y+ QI+ H+ V VGE S MS Y+GG ++SLDI+EVVN N+
Sbjct 327 EFLQKWKEITQSGNKDYKDQIEKHWNVSVGEAYSEMSLYLGGTTASLDINEVVNNNITGS 386
Query 404 DVASEAVIAGKGVGSSQGSEKFEARD-WGVLMCIYHNVPLLDYVSSAPDPQFFVTQNTDL 462
+ A IAGKGV G F+A + +G++MCIYH++PLLDY + +P F +TD
Sbjct 387 NAAD---IAGKGVVVGNGRISFDAGERYGLIMCIYHSLPLLDYTTDLVNPAFTKINSTDF 443
Query 463 PIPELDSIGMQSVPLAMYTNSDGELVSGFVSPDYTMGYLPRYFSWKTSYDYVLGAFTTTE 522
IPE D +GM+SVPL N L S + +GY PRY S+KT D +GAF TT
Sbjct 444 AIPEFDRVGMESVPLVSLMN---PLQSSYNVGSSILGYAPRYISYKTDVDSSVGAFKTTL 500
Query 523 KEWVAPISSALWKNMLSTITVRNPQ----FTYNFFKVNPSVLDSIFQVNADSKWDTDPFL 578
K WV + N L+ N Y FKVNP+ +D +F V A + DTD FL
Sbjct 501 KSWVMSYDNQSVINQLNYQDDPNNSPGTLVNYTNFKVNPNCVDPLFAVAASNSIDTDQFL 560
Query 579 INCAFDVKVVRNLDYSGMPY 598
+ FDVKVVRNLD G+PY
Sbjct 561 CSSFFDVKVVRNLDTDGLPY 580
>gi|490418709|ref|WP_004291032.1| hypothetical protein [Bacteroides eggerthii]
gi|217986636|gb|EEC52970.1| putative capsid protein (F protein) [Bacteroides eggerthii DSM
20697]
Length=578
Score = 400 bits (1027), Expect = 5e-128, Method: Compositional matrix adjust.
Identities = 245/624 (39%), Positives = 337/624 (54%), Gaps = 75/624 (12%)
Query 2 SLFSLKDIRNHPRRSAFDLSSKVAFSAKSGELLPIKWYFTMPGDKFTLKRQHFTRTQPVN 61
++ SLK IRN P R+ FDLS K F+AK+GELLP+ +PGD F + + FTRTQPVN
Sbjct 3 NIMSLKSIRNKPSRNGFDLSFKKNFTAKAGELLPVMVKEVLPGDTFKINLKAFTRTQPVN 62
Query 62 TSAYTRIREYYDWFWVPLHLLWRHAPEVISQMQSNVQHAGS--QTSSLTLGNYLPTISSS 119
T+A+ RIREYYD+F+VP LLW A V++QM N QHA S T + L +P ++S
Sbjct 63 TAAFARIREYYDFFFVPYDLLWNKANTVLTQMYDNPQHAVSIDPTRNFVLSGEMPYMTSE 122
Query 120 QLSAVCSRLFG-------KKNYFGYDRSDLSYKLMQYLRVGNSGQVSVNFGTSLPASDTS 172
+++ + L K NYFGY+RS S KL++YL GN +D
Sbjct 123 AIASYINALSTASALADYKSNYFGYNRSKSSVKLLEYLGYGNYESF---------LTDDW 173
Query 173 YTQAYRFNLDLSLFPFLAYKKFCQDYFRYSQWQDSSPYLWNIDYFTGvsshlfsslpvss 232
T NL+ ++F LAY+K D++R SQW+ SP +N+DY G S +L ++
Sbjct 174 NTAPLMANLNHNIFGLLAYQKIYSDFYRDSQWERVSPSTFNVDYLDGSSMNLDNAYSTE- 232
Query 233 DPYWNNNTLFDLEYCNWNKDMFMGVFPDTQFGDVATIGITSDSPESSLQLKAWasgspss 292
++ N FDL YCNW KD+F GV P Q+G+ A IT D L L
Sbjct 233 --FYQNYNFFDLRYCNWQKDLFHGVLPHQQYGETAVASITPDV-TGKLTLS--------- 280
Query 293 kapvvvgaaasspNFTIRAESGNMNPANILGVDTSSL---SLAGSFDVLALRRGEALQRW 349
NF+ S P G T +L G +L LR+ E LQ+W
Sbjct 281 -------------NFSTVGTS----PTTASGTATKNLPAFDTVGDLSILVLRQAEFLQKW 323
Query 350 KEISLNVPQNYRAQIKAHFGVDVGENMSGMSTYVGGDSSSLDISEVVNTNLQSGDVASEA 409
KEI+ + ++Y+ Q++ H+GV VG+ S + TY+GG SSS+DI+EV+NTN+ +G A++
Sbjct 324 KEITQSGNKDYKDQLEKHWGVSVGDGFSELCTYLGGVSSSIDINEVINTNI-TGSAAAD- 381
Query 410 VIAGKGVGSSQGSEKFEARD-WGVLMCIYHNVPLLDYVSSAPDPQFFVTQNTDLPIPELD 468
IAGKGVG + G F + +G++MCIYH +PLLDY + DP F +TD IPE D
Sbjct 382 -IAGKGVGVANGEINFNSNGRYGLIMCIYHCLPLLDYTTDMLDPAFLKVNSTDYAIPEFD 440
Query 469 SIGMQSVPLAMYTNSDGELVSGFVSPDYTMGYLPRYFSWKTSYDYVLGAFTTTEKEWVAP 528
+GMQS+PL N L S + +GY+PRY +KTS D +G F T WV
Sbjct 441 RVGMQSMPLVQLMN---PLRSFANASGLVLGYVPRYIDYKTSVDQSVGGFKRTLNSWVIS 497
Query 529 ISSALWKNMLSTITVRN--------------PQFTYNFFKVNPSVLDSIFQVNADSKWDT 574
+ ++L +T+ N + FFKVNP LD IF V A +T
Sbjct 498 YGNI---SVLKQVTLPNDAPPIEPSEPVPSVAPMNFTFFKVNPDCLDPIFAVQAGDDTNT 554
Query 575 DPFLINCAFDVKVVRNLDYSGMPY 598
D FL + FD+K VRNLD G+PY
Sbjct 555 DQFLCSSFFDIKAVRNLDTDGLPY 578
>gi|575094354|emb|CDL65742.1| unnamed protein product [uncultured bacterium]
Length=615
Score = 394 bits (1012), Expect = 3e-125, Method: Compositional matrix adjust.
Identities = 243/638 (38%), Positives = 351/638 (55%), Gaps = 68/638 (11%)
Query 5 SLKDIRNHPRRSAFDLSSKVAFSAKSGELLPIKWYFTMPGDKFTLKRQHFTRTQPVNTSA 64
S+ DI+N P R+ FDLS K F+AK+GELLP+ +PGD F + + FTRTQP+NTSA
Sbjct 2 SMADIKNRPSRNGFDLSFKKNFTAKAGELLPVMTKVVLPGDSFNINLRSFTRTQPLNTSA 61
Query 65 YTRIREYYDWFWVPLHLLWRHAPEVISQMQSNVQHAGSQT--SSLTLGNYLPTISSSQLS 122
+ R+REYYD+++VP +W I+QM +NVQHA T + L +P +S Q++
Sbjct 62 FARMREYYDFYFVPFEQMWNKFDSCITQMNANVQHASGPTLDDNTPLSGRMPYFTSEQIA 121
Query 123 AVCS--RLFGKKNYFGYDRSDLSYKLMQYLRVGNSGQVSVNFGTSLPASDTSYTQAYRFN 180
+ +KN FG++RS L+ KL+QYL G+ + + ++T + +N
Sbjct 122 DYLNDQATAARKNPFGFNRSTLTCKLLQYLGYGD-------YNSFDSETNTWSAKPLLYN 174
Query 181 LDLSLFPFLAYKKFCQDYFRYSQWQDSSPYLWNIDYFTGvsshlfsslpvssDPYWNNNT 240
L+LS FP LAY+K D++RY+QW+ ++P +N+DY G S + SD +N
Sbjct 175 LELSPFPLLAYQKIYSDFYRYTQWEKTNPSTFNLDYIKGTSDLQMDLTGLPSD----DNN 230
Query 241 LFDLEYCNWNKDMFMGVFPDTQFGDVATIGITSDSP-ESSLQLKAWasgspsskapvvvg 299
FD+ YCN+ KDMF GV P VA G S P L + + P K
Sbjct 231 FFDIRYCNYQKDMFHGVLP------VAQYGSASVVPINGQLNVISNGDSGPIFKTSTPDP 284
Query 300 aaasspNFTIRAESGNMNPANILGVDTSSLSLAGSFD----------------------- 336
+ T+ G N + GV S+L++ S D
Sbjct 285 GTPGTSYVTVGGNIGVDNRS--FGVSGSTLNVGKSADPSGYGFPSNASTRSLLWENPNLI 342
Query 337 ----------VLALRRGEALQRWKEISLNVPQNYRAQIKAHFGVDVGENMSGMSTYVGGD 386
+LALR+ E LQ+WKE+S++ ++Y++QI+ H+G+ V + +S + Y+GG
Sbjct 343 IENNQGFYVPILALRQAEFLQKWKEVSVSGEEDYKSQIEKHWGIKVSDFLSHQARYLGGC 402
Query 387 SSSLDISEVVNTNLQSGDVASEAVIAGKGVGSSQGSEKFEAR-DWGVLMCIYHNVPLLDY 445
++SLDI+EV+N N+ +GD A++ IAGKG + GS +FE++ ++G++MCIYH +P++DY
Sbjct 403 ATSLDINEVINNNI-TGDNAAD--IAGKGTFTGNGSIRFESKGEYGIIMCIYHVLPIVDY 459
Query 446 VSSAPDPQFFVTQNTDLPIPELDSIGMQSVPLAMYTNSDGELVSGFVSPDYTMGYLPRYF 505
V S D + T PIPELD IGM+SVPL N E S S D +GY PRY
Sbjct 460 VGSGVDHSCTLVDATSFPIPELDQIGMESVPLVRAMNPVKE--SDTPSADTFLGYAPRYI 517
Query 506 SWKTSYDYVLGAFTTTEKEWVAPI-----SSALWKNMLSTITVRNPQFTYNFFKVNPSVL 560
WKTS D +G F + + W P+ +SA N S V FFKVNPS++
Sbjct 518 DWKTSVDRSVGDFADSLRTWCLPVGDKELTSANSLNFPSNPNVEPDSIAAGFFKVNPSIV 577
Query 561 DSIFQVNADSKWDTDPFLINCAFDVKVVRNLDYSGMPY 598
D +F V ADS TD FL + FDVKVVRNLD +G+PY
Sbjct 578 DPLFAVVADSTVKTDEFLCSSFFDVKVVRNLDVNGLPY 615
>gi|547226430|ref|WP_021963493.1| putative uncharacterized protein [Prevotella sp. CAG:1185]
gi|524103382|emb|CCY83994.1| putative uncharacterized protein [Prevotella sp. CAG:1185]
Length=573
Score = 390 bits (1002), Expect = 2e-124, Method: Compositional matrix adjust.
Identities = 243/617 (39%), Positives = 345/617 (56%), Gaps = 66/617 (11%)
Query 2 SLFSLKDIRNHPRRSAFDLSSKVAFSAKSGELLPIKWYFTMPGDKFTLKRQHFTRTQPVN 61
S+ SL ++N +R+ FDLS K AF+AK GELLPI PGDKF ++ Q FTRTQPVN
Sbjct 3 SVMSLTALKNSVKRNGFDLSFKNAFTAKVGELLPIMCKEVYPGDKFNIRGQAFTRTQPVN 62
Query 62 TSAYTRIREYYDWFWVPLHLLWRHAPEVISQMQSNVQHAGSQTSSLTLG----------- 110
++AY+R+REYYD+++VP LLW AP + M + HA SS+ L
Sbjct 63 SAAYSRLREYYDFYFVPYRLLWNMAPTFFTNM-PDPHHAADLVSSVNLSQRHPWFTFFDI 121
Query 111 -NYLPTISSSQLSAVCSRLFGKKNYFGYDRSDLSYKLMQYLRVGNSGQVSVNFGTSLPAS 169
YL ++S LS + +KN+FG+ R +LS KL+ YL G ++ + S
Sbjct 122 MEYLGNLNS--LSGAYEKY--QKNFFGFSRVELSVKLLNYLNYG----FGKDYESVKVPS 173
Query 170 DTSYTQAYRFNLDLSLFPFLAYKKFCQDYFRYSQWQDSSPYLWNIDYFTGvsshlfsslp 229
D+ ++ LS FP LAY+K C+DYFR QWQ ++PY +N+DY G SS +
Sbjct 174 DSD-------DIVLSPFPLLAYQKICEDYFRDDQWQSAAPYRYNLDYLYGKSSGFHIPMS 226
Query 230 vssDPYWNNNTLFDLEYCNWNKDMFMGVFPDTQFGDVAT----IGITSDSPESSLQLKAW 285
++ + N T+FDL YCN+ KD F G+ P Q+GDV+ G SSL
Sbjct 227 SFTNDAFKNPTMFDLNYCNFQKDYFTGMLPRAQYGDVSVASPIFGDLDIGDSSSLTFA-- 284
Query 286 asgspsskapvvvgaaasspNFTIRAESGNMNPANILGVDTSSLSLAGSFDVLALRRGEA 345
+ + N + +L V+ +S + AG VLALR+ E
Sbjct 285 ----------------------SAPQQGANTIQSGVLVVNNNSNTTAG-LSVLALRQAEC 321
Query 346 LQRWKEISLNVPQNYRAQIKAHFGVDVGENMSGMSTYVGGDSSSLDISEVVNTNLQSGDV 405
LQ+W+EI+ + +Y+ Q++ HF V +SG Y+GG +S+LDISEVVNTNL +GD
Sbjct 322 LQKWREIAQSGKMDYQTQMQKHFNVSPSATLSGHCKYLGGWTSNLDISEVVNTNL-TGD- 379
Query 406 ASEAVIAGKGVGSSQGSE-KFEARDWGVLMCIYHNVPLLDYVSSAPDPQFFVTQNTDLPI 464
++A I GKG G+ G++ FE+ + G++MCIYH +PLLD+ + Q F T TD I
Sbjct 380 -NQADIQGKGTGTLNGNKVDFESSEHGIIMCIYHCLPLLDWSINRIARQNFKTTFTDYAI 438
Query 465 PELDSIGMQSVPLAMYTNSDGELVSGFVSPDYTMGYLPRYFSWKTSYDYVLGAFTTTEKE 524
PE DS+GMQ + + +L S S + MGY+PRY KTS D + G+F T
Sbjct 439 PEFDSVGMQQLYPSEMIFGLEDLPSDPSSIN--MGYVPRYADLKTSIDEIHGSFIDTLVS 496
Query 525 WVAPISSA---LWKNMLSTITVRNPQFTYNFFKVNPSVLDSIFQVNADSKWDTDPFLINC 581
WV+P++ + ++ + TYNFFKVNP ++D+IF V ADS +TD LIN
Sbjct 497 WVSPLTDSYISAYRQACKDAGFSDITMTYNFFKVNPHIVDNIFGVKADSTINTDQLLINS 556
Query 582 AFDVKVVRNLDYSGMPY 598
FD+K VRN DY+G+PY
Sbjct 557 YFDIKAVRNFDYNGLPY 573
>gi|494822885|ref|WP_007558293.1| hypothetical protein [Bacteroides plebeius]
gi|198272099|gb|EDY96368.1| putative capsid protein (F protein) [Bacteroides plebeius DSM
17135]
Length=613
Score = 351 bits (900), Expect = 1e-108, Method: Compositional matrix adjust.
Identities = 220/621 (35%), Positives = 338/621 (54%), Gaps = 41/621 (7%)
Query 2 SLFSLKDIRNHPRRSAFDLSSKVAFSAKSGELLPIKWYFTMPGDKFTLKRQHFTRTQPVN 61
++ S+K +RN P R+ +DL+ K+ F+AK+G L+P+ W +P D + F RTQP+N
Sbjct 10 NIMSMKSVRNKPTRAGYDLTQKINFTAKAGSLIPVWWTPVLPFDDLNATVKSFVRTQPLN 69
Query 62 TSAYTRIREYYDWFWVPLHLLWRHAPEVISQMQSNVQHAGSQT--SSLTLGNYLPTISSS 119
T+A+ R+R Y+D+++VP +W P I+QM++N+ HA ++ L + LP ++
Sbjct 70 TAAFARMRGYFDFYFVPFRQMWNKFPTAITQMRTNLLHASGPVLADNVPLSDELPYFTAE 129
Query 120 QLSAVCSRLFGKKNYFGYDRSDLSYKLMQYLRVGNSGQVSVNFGTSLPASDTSYTQAYRF 179
Q++ L KN FGY R+ L +++YL G+ V A T T+
Sbjct 130 QVADYIVSLADSKNQFGYYRAWLVCIILEYLGYGDFYPYIVEAAGGEGA--TWATRPMLN 187
Query 180 NLDLSLFPFLAYKKFCQDYFRYSQWQDSSPYLWNIDYFTGvsshlfsslpvssDPYWNNN 239
NL S FP AY+K D+ RY+QW+ S+P +NIDY +G S L + + + ++
Sbjct 188 NLKFSPFPLFAYQKIYADFNRYTQWERSNPSTFNIDYISG--SADSLQLDFTVEGFKDSF 245
Query 240 TLFDLEYCNWNKDMFMGVFPDTQFGDVATIGITSDSPESSLQLKAWasgspsskapvvvg 299
LFD+ Y NW +D+ G P Q+G+ + + ++ ++ +P + G
Sbjct 246 NLFDMRYSNWQRDLLHGTIPQAQYGEASAVPVSG-------SMQVVEGPTPPAFTTGQDG 298
Query 300 aaasspNFTIRAESGNMNPANILGVD--------TSSLSLAG--SFDV--LALRRGEALQ 347
A + N TI+ SG + +G S L + G SF V LALRR EA Q
Sbjct 299 VAFLNGNVTIQGSSGYLQAQTSVGESRILRFNNTNSGLIVEGDSSFGVSILALRRAEAAQ 358
Query 348 RWKEISLNVPQNYRAQIKAHFGVDVGENMSGMSTYVGGDSSSLDISEVVNTNLQSGDVAS 407
+WKE++L ++Y +QI+AH+G V + S M ++G + L I+EVVN N+ +G+ A+
Sbjct 359 KWKEVALASEEDYPSQIEAHWGQSVNKAYSDMCQWLGSINIDLSINEVVNNNI-TGENAA 417
Query 408 EAVIAGKGVGSSQGSEKFE-ARDWGVLMCIYHNVPLLDYVSSAPDPQFFVTQNTDLPIPE 466
+ IAGKG S GS F +G++MC++H +P LDY++SAP +T D PIPE
Sbjct 418 D--IAGKGTMSGNGSINFNVGGQYGIVMCVFHVLPQLDYITSAPHFGTTLTNVLDFPIPE 475
Query 467 LDSIGMQSVPLAMYTN----SDGELVSGFVSPDYTMGYLPRYFSWKTSYDYVLGAFTTTE 522
D IGM+ VP+ N DG+ VSP+ GY P+Y++WKT+ D +G F +
Sbjct 476 FDKIGMEQVPVIRGLNPVKPKDGDFK---VSPNLYFGYAPQYYNWKTTLDKSMGEFRRSL 532
Query 523 KEWVAPISSALWKNMLSTITVRNPQFTYN-----FFKVNPSVLDSIFQVNADSKWDTDPF 577
K W+ P S NP + FFKV+PSVLD++F V A+S +TD F
Sbjct 533 KTWIIPFDDEALLAADSVDFPDNPNVEADSVKAGFFKVSPSVLDNLFAVKANSDLNTDQF 592
Query 578 LINCAFDVKVVRNLDYSGMPY 598
L + FDV VVR+LD +G+PY
Sbjct 593 LCSTLFDVNVVRSLDPNGLPY 613
>gi|575094321|emb|CDL65708.1| unnamed protein product [uncultured bacterium]
Length=642
Score = 241 bits (614), Expect = 7e-67, Method: Compositional matrix adjust.
Identities = 198/645 (31%), Positives = 294/645 (46%), Gaps = 58/645 (9%)
Query 2 SLFSLKDIRNHPRRSAFDLSSKVAFSAKSGELLPIKWYFTMPGDKFTLKRQHFTRTQPVN 61
++ L ++N P R++FDLS + F+AK GELLP PGD + +FTRT P+
Sbjct 6 NIMGLHGLKNKPSRNSFDLSHRNMFTAKVGELLPCFVQELNPGDSVKVSSSYFTRTAPLQ 65
Query 62 TSAYTRIREYYDWFWVPLHLLWRHAPEVISQMQSNVQHAG-SQTSSLTLGNY-----LPT 115
++A+TR+RE +F+VP LW++ + M N S+ +S +GN +P
Sbjct 66 SNAFTRLRENVQYFFVPYSALWKYFDSQVLNMTKNANGGDISRIASSLVGNQKVTTQMPC 125
Query 116 ISSSQLSAVCSRLFGKKNYFGYD------------RSDLSYKLMQYLRVGNSGQVSVNFG 163
++ L A + F ++ G D R S KL+Q L GN + NF
Sbjct 126 VNYKTLHAYLLK-FINRSTVGSDGSVGPEFNRGCYRHAESAKLLQLLGYGNFPEQFANFK 184
Query 164 TSLPASDTSYTQ----AYRFNLDLSLFPFLAYKKFCQDYFRYSQWQDSSPYLWNIDYFTG 219
+ + S Y + LS+F LAY K C D++ Y QWQ + L N+DY T
Sbjct 185 VNNDKHNQSGQNFKDVTYNNSPYLSIFRLLAYHKICNDHYLYRQWQPYNASLCNVDYLTP 244
Query 220 vsshlfsslpvssDP-----YWNNNTLFDLEYCNWNKDMFMGVFPDTQFGDVATIGITSD 274
SS L S L D+ + N D F GV P +QFG + + +
Sbjct 245 NSSSLLSIDDALLSIPDDSIKAEKLNLLDMRFSNLPLDYFTGVLPTSQFGSESVVNLNLG 304
Query 275 SPESSLQLKAWasgspsskapvvvgaaasspNFTIRAESGNMNPANILGVDTS------- 327
+ S L S + +GN+ N G S
Sbjct 305 NASGSAVLNGTTSKDSGRWRTTTGEWEMEQR--VASSANGNLKLDNSNGTFISHDHTFSG 362
Query 328 ----SLSLAGSFDVLALRRGEALQRWKEISLNVPQNYRAQIKAHFGVDVGENMSGMSTYV 383
+ SL+G+ ++ALR A Q++KEI L ++++Q++AHFG+ E S ++
Sbjct 363 NVAINTSLSGNLSIIALRNALAAQKYKEIQLANDVDFQSQVEAHFGIKPDEKNEN-SLFI 421
Query 384 GGDSSSLDISEVVNTNLQSGDVASEAVIAGKGVGSSQGSEKFEARDWGVLMCIYHNVPLL 443
GG SS ++I+E +N NL SGD + A +G GS+ S KF A+ +GV++ IY P+L
Sbjct 422 GGSSSMININEQINQNL-SGDNKATYGAAPQGNGSA--SIKFTAKTYGVVIGIYRCTPVL 478
Query 444 DYVSSAPDPQFFVTQNTDLPIPELDSIGMQ-------SVPLAMYTNSDGELVSGFVSPDY 496
D+ D F T +D IPE+DSIGMQ + P V SPD
Sbjct 479 DFAHLGIDRTLFKTDASDFVIPEMDSIGMQQTFRCEVAAPAPYNDEFKAFRVGDGSSPDM 538
Query 497 --TMGYLPRYFSWKTSYDYVLGAFTTTEKEWVAPIS-SALWKNMLSTITVRNPQFTYNFF 553
T GY PRY +KTSYD GAF + K WV I+ A+ N+ +T N N F
Sbjct 539 SETYGYAPRYSEFKTSYDRYNGAFCHSLKSWVTGINFDAIQNNVWNTWAGINAP---NMF 595
Query 554 KVNPSVLDSIFQVNADSKWDTDPFLINCAFDVKVVRNLDYSGMPY 598
P ++ ++F V++ + D D + RNL G+PY
Sbjct 596 ACRPDIVKNLFLVSSTNNSDDDQLYVGMVNMCYATRNLSRYGLPY 640
>gi|494308783|ref|WP_007173938.1| hypothetical protein [Prevotella bergensis]
gi|270333035|gb|EFA43821.1| putative capsid protein (F protein) [Prevotella bergensis DSM
17361]
Length=553
Score = 200 bits (508), Expect = 8e-53, Method: Compositional matrix adjust.
Identities = 174/619 (28%), Positives = 269/619 (43%), Gaps = 92/619 (15%)
Query 1 MSLFSLKDIRNHPRRSAFDLSSKVAFSAKSGELLPIKWYFTMPGDKFTLKRQHFTRTQPV 60
+S+ +K R + R+AFDLS + F+A +G LLP+ +P D + Q F RT P+
Sbjct 3 VSIPKIKATRPNRNRNAFDLSQRHLFTAHAGMLLPVLNLDLIPHDHVEINAQDFMRTLPM 62
Query 61 NTSAYTRIREYYDWFWVPLHLLWRHAPEVISQMQSNVQHAGSQTSSLTLGNYLPTISSSQ 120
NT+A+ +R Y++F+VP H LW + I+ M N H+ S S+ G +
Sbjct 63 NTAAFASMRGVYEFFFVPYHQLWAQFDQFITGM--NDFHS-SANKSIQGGTSPLQVPYFN 119
Query 121 LSAVCSRLFGKKNYFGYDRSDLSYKL----MQYLRVGNSGQVSVNFGTSLPASDTSYTQA 176
+ +V + L K DL YK + L + G+ +FGT+ P + +
Sbjct 120 VDSVFNSLNTGKESGSGSTDDLQYKFKYGAFRLLDLLGYGRKFDSFGTAYPDNVSGLKNN 179
Query 177 YRFNLDLSLFPFLAYKKFCQDYFRYSQWQDSSPYLWNIDYFTGvsshlfsslpvssDPYW 236
+N S+F LAY K QDY+R S +++ +N D F G
Sbjct 180 LDYNC--SVFRILAYNKIYQDYYRNSNYENFDTDSFNFDKFKGGLVDAKVVA-------- 229
Query 237 NNNTLFDLEYCNWNKDMFMGVFPD------TQFGDVATIGITSDSPESSLQLKAWasgsp 290
LF L Y N D F + T F DV I I +P
Sbjct 230 ---DLFKLRYRNAQTDYFTNLRQSQLFSFTTAFEDVDNINI---APRD------------ 271
Query 291 sskapvvvgaaasspNFTIRAESGNMNPANILGVDTSSLSLAGSFDVLALRRGEALQRWK 350
++++ N N GVDT S G F V +LR A+ +
Sbjct 272 -----------------YVKSDGSNFTRVN-FGVDTDSSE--GDFSVSSLRAAFAVDKLL 311
Query 351 EISLNVPQNYRAQIKAHFGVDVGENMSGMSTYVGGDSSSLDISEVVNTNLQSGDVASE-- 408
+++ + ++ Q++AH+GV++ ++ G Y+GG S + +S+V T SG A+E
Sbjct 312 SVTMRAGKTFQDQMRAHYGVEIPDSRDGRVNYLGGFDSDMQVSDVTQT---SGTTATEYK 368
Query 409 ------AVIAGKGVGSSQGSEKFEARDWGVLMCIYHNVPLLDYVSSAPDPQFFVTQNTDL 462
+AGKG GS +G F+A++ GVLMCIY VP + Y + DP D
Sbjct 369 PEAGYLGRVAGKGTGSGRGRIVFDAKEHGVLMCIYSLVPQIQYDCTRLDPMVDKLDRFDY 428
Query 463 PIPELDSIGMQSVPLAMYTNSDGELVSGFVSPD---YTMGYLPRYFSWKTSYDYVLGAFT 519
PE +++GMQ PL + +S F + D +GY PRY +KT+ D G F
Sbjct 429 FTPEFENLGMQ--PL------NSSYISSFCTTDPKNPVLGYQPRYSEYKTALDVNHGQFA 480
Query 520 TTEKEWVAPISS-ALWKNMLSTITVRNPQFTYNFFKVNPSVLDSIFQVNADSKWDTDPFL 578
++ +S W PQ FK++P L+SIF V+ + D
Sbjct 481 QSDALSSWSVSRFRRWTTF--------PQLEIADFKIDPGCLNSIFPVDYNGTEANDCVY 532
Query 579 INCAFDVKVVRNLDYSGMP 597
C F++ V ++ GMP
Sbjct 533 GGCNFNIVKVSDMSVDGMP 551
>gi|496521299|ref|WP_009229582.1| capsid protein [Prevotella sp. oral taxon 317]
gi|288330570|gb|EFC69154.1| putative capsid protein (F protein) [Prevotella sp. oral taxon
317 str. F0108]
Length=541
Score = 199 bits (506), Expect = 1e-52, Method: Compositional matrix adjust.
Identities = 184/615 (30%), Positives = 269/615 (44%), Gaps = 94/615 (15%)
Query 1 MSLFSLKDIR----NHPRRSAFDLSSKVAFSAKSGELLPIKWYFTMPGDKFTLKRQHFTR 56
MSL + I+ N PR SAFDLS K ++A +G LLP+ M D ++ Q F R
Sbjct 1 MSLKKVPQIKPSRANRPR-SAFDLSQKHLYTAPAGALLPVLSVDLMFHDHIRIQAQDFMR 59
Query 57 TQPVNTSAYTRIREYYDWFWVPLHLLWRHAPEVISQMQSNVQHAGSQTSSLTLGNYLPTI 116
T P+N++A+ +R Y++F+VP LW + I+ M + S SS L ++
Sbjct 60 TMPMNSAAFISMRGVYEFFFVPYSQLWHPYDQFITSMN---DYRSSVVSSAAGDKALDSV 116
Query 117 SSSQLSAVCS--RLFGKKNYFGYDRSDLSYKLMQYLRVGNSGQVSVNFGTSLPASDTSYT 174
+ +L+ + R K+ FGY S+ S +LM L +G + +S T
Sbjct 117 PNVKLADMYKFVRERTDKDIFGYPHSNNSCRLMDLL----------GYGKPITSSKTPVP 166
Query 175 QAYRFNLDLSLFPFLAYKKFCQDYFRYSQWQDSSPYLWNIDYFTGvsshlfsslpvssDP 234
Y N++ LF LAY K DY+R + ++ Y +NID+ G
Sbjct 167 LLYTGNVN--LFRLLAYNKIYSDYYRNTTYEGVDVYSFNIDHKKGTFVPTADEF------ 218
Query 235 YWNNNTLFDLEYCNWNKDMFMGVFPDTQFGDVATIGITSDSPESSLQLKAWasgspsska 294
+L Y N D + + P F TIG SDS S LQL
Sbjct 219 ----KKYLNLHYRNAPLDFYTNLRPTPLF----TIG--SDSFSSVLQLS----------- 257
Query 295 pvvvgaaasspNFTIRAESGNMNPANILGVDTSSLSLAGSFDVLALRRGEALQRWKEISL 354
S F+ S +N A+ +V A+R AL + IS+
Sbjct 258 -----DPTGSAGFSADGNSAKLNMAS-----------PDVLNVSAIRSAFALDKLLSISM 301
Query 355 NVPQNYRAQIKAHFGVDVGENMSGMSTYVGGDSSSLDISEVVNTNLQSGDVASE------ 408
+ Y QI+AHFGV V E G Y+GG S++ + +V T+ + SE
Sbjct 302 RAGKTYAEQIEAHFGVTVSEGRDGQVYYLGGFDSNVQVGDVTQTSGTTNPNVSEVGNAKL 361
Query 409 ----AVIAGKGVGSSQGSEKFEARDWGVLMCIYHNVPLLDYVSSAPDPQFFVTQNT--DL 462
I GKG GS G +F+A++ GVLMCIY VP + Y DP FV + T D
Sbjct 362 AGYLGKITGKGTGSGYGEIQFDAKEPGVLMCIYSVVPAMQYDCMRLDP--FVAKQTRGDY 419
Query 463 PIPELDSIGMQSVPLAMYTNSDGELVSGFVSPDYTMGYLPRYFSWKTSYDYVLGAFTTTE 522
IPE +++GMQ + A VS + D + G+ PRY +KT++D G F E
Sbjct 420 FIPEFENLGMQPIVPA--------FVSLNRAKDNSYGWQPRYSEYKTAFDINHGQFANGE 471
Query 523 KEWVAPISSALWKNMLSTITVRNPQFTYNFFKVNPSVLDSIFQVNADSKWDTDPFLINCA 582
I+ A + L+T V K+NP LDS+F VN + TD
Sbjct 472 PLSYWSIARARGSDTLNTFNVAA-------LKINPHWLDSVFAVNYNGTEVTDCMFGYAH 524
Query 583 FDVKVVRNLDYSGMP 597
F+++ V ++ GMP
Sbjct 525 FNIEKVSDMTEDGMP 539
>gi|494306153|ref|WP_007173049.1| hypothetical protein [Prevotella bergensis]
gi|270333881|gb|EFA44667.1| putative capsid protein (F protein) [Prevotella bergensis DSM
17361]
Length=519
Score = 173 bits (438), Expect = 1e-43, Method: Compositional matrix adjust.
Identities = 156/580 (27%), Positives = 253/580 (44%), Gaps = 79/580 (14%)
Query 33 LLPIKWYFTMPGDKFTLKRQHFTRTQPVNTSAYTRIREYYDWFWVPLHLLWRHAPEVISQ 92
LLP+ +P D + Q F RT P+NT+A+ +R Y++F+VP H LW + I+
Sbjct 2 LLPVLNLDLIPHDHVEINAQDFMRTLPMNTAAFASMRGVYEFFFVPYHQLWAQFDQFITG 61
Query 93 MQSNVQHAGSQTSSLTLGNYLPTISSSQLSAVCSRLFGKKNYFGYDRSDLSYKL----MQ 148
M N H+ S S+ G + L +V + + + + + DL Y+ +
Sbjct 62 M--NDFHS-SANKSIQGGTSPLQVPYFNLESVFKNIIERDSTPSF-QDDLQYRFKYGAFR 117
Query 149 YLRVGNSGQVSVNFGTSLPASDTSYTQAYRFNLDLSLFPFLAYKKFCQDYFRYSQWQDSS 208
L + G+ +FGT+ P + + +N S+F LAY K QDY+R S +++
Sbjct 118 LLDLLGYGRKFDSFGTAYPDNVSGLKNNLDYNC--SVFRVLAYNKIYQDYYRNSNYENFD 175
Query 209 PYLWNIDYFTGvsshlfsslpvssDPYWNNNTLFDLEYCNWNKDMFMGVFPDTQFGDVAT 268
+N D F G LF L Y N D F + F +
Sbjct 176 TDSFNFDKFKGGLVDAKVVA-----------DLFKLRYRNAQTDYFTNLRQSQLFTFIPE 224
Query 269 IGITSDSPESSLQLKAWasgspsskapvvvgaaasspNFTIRAESGNMNPANILGVDTSS 328
SD + +A S S+ + NF + ++
Sbjct 225 F---SDDEHLNFDRDQYADQSKSNFTQL---------NFPVDVDNN-------------- 258
Query 329 LSLAGSFDVLALRRGEALQRWKEISLNVPQNYRAQIKAHFGVDVGENMSGMSTYVGGDSS 388
G F V +LR A+ + +++ + ++ Q++AH+GV++ ++ G Y+GG S
Sbjct 259 ---LGYFSVSSLRSAFAVDKLLSVTMRAGKTFQDQMRAHYGVEIPDSRDGRVNYLGGFDS 315
Query 389 SLDISEVVNTNLQSGDVASE--------AVIAGKGVGSSQGSEKFEARDWGVLMCIYHNV 440
L +S+V T SG A+E IAGKG GS +G F+A++ GVLMCIY V
Sbjct 316 DLQVSDVTQT---SGTTATEYKPEAGYLGRIAGKGTGSGRGRIVFDAKEHGVLMCIYSLV 372
Query 441 PLLDYVSSAPDPQFFVTQNTDLPIPELDSIGMQSVPLAMYTNSDGELVSGFVSPD---YT 497
P + Y + DP D PE +++GMQ PL + +S F +PD
Sbjct 373 PQIQYDCTRLDPMVDKLDRFDFFTPEFENLGMQ--PL------NSSYISSFCTPDPKNPV 424
Query 498 MGYLPRYFSWKTSYDYVLGAFTTTEKEWVAPISSALWKNMLSTITVRNPQFTYNFFKVNP 557
+GY PRY +KT+ D G F + + ++ S + ++ + PQ FK++P
Sbjct 425 LGYQPRYSEYKTALDINHGQF--AQNDALSSWSVSRFRRWTTF-----PQLEIADFKIDP 477
Query 558 SVLDSIFQVNADSKWDTDPFLINCAFDVKVVRNLDYSGMP 597
L+S+F V + TD C F++ V ++ GMP
Sbjct 478 GCLNSVFPVEFNGTESTDCVFGGCNFNIVKVSDMSVDGMP 517
>gi|517172762|ref|WP_018361580.1| hypothetical protein [Prevotella nanceiensis]
Length=568
Score = 157 bits (398), Expect = 4e-38, Method: Compositional matrix adjust.
Identities = 150/598 (25%), Positives = 258/598 (43%), Gaps = 64/598 (11%)
Query 15 RSAFDLSSKVAFSAKSGELLPIKWYFTMPGDKFTLKRQHFTRTQPVNTSAYTRIREYYDW 74
R+AFD+S + F+A +G LLP+ +P D + F RT P+N++A+ +R Y++
Sbjct 18 RNAFDISQRHLFTAPAGALLPVLSLDLLPHDHVEINASDFMRTLPMNSAAFMSMRGVYEF 77
Query 75 FWVPLHLLWRHAPEVISQM---QSNVQHAGSQTSSLTLGNYLPTISSSQLSAVC--SRLF 129
++VP LW + I+ M +S+ +A G P+ S + + +
Sbjct 78 YFVPYKQLWSGFDQFITGMSDYKSSFMYAFK-------GKTPPSCVSFDVQKLVDWCKTN 130
Query 130 GKKNYFGYDRSDLSYKLMQYLRVGNSGQVSVNFGTSLPASDTSYTQAYRFNLDLSLFPFL 189
K+ G+D++ Y+++ L G + +P ++ + T + + F L
Sbjct 131 TAKDIHGFDKNKGVYRILDLLGYGKYANSA-----GVPYTNPTSTTMGK----CTPFRGL 181
Query 190 AYKKFCQDYFRYSQWQDSSPYLWNIDYFTGvsshlfsslpvssDPYWNNNTLFDLEYCNW 249
AY+K D++R + +++ +N+D F G + D W F L Y N
Sbjct 182 AYQKIYNDFYRNTTYEEYQLESFNVDMFYGSGKVKETIPNEPWDYDW-----FTLRYRNA 236
Query 250 NKDMFMGVFPDTQFGDVATIGITSDSPESSLQLKAWasgspsskapvvvgaaasspNFTI 309
KD+ V P F I +P+ + V G + I
Sbjct 237 QKDLLTNVRPTPLFS------IDDFNPQF---FTGGSDIVMEKGPNVTGGTHEYRDSVVI 287
Query 310 RAESGNMNPANILGVDTSSLSLAGSFDVLALRRGEALQRWKEISLNVPQNYRAQIKAHFG 369
++ N GVD+ ++ V +R AL++ +++ + Y+ Q++AHFG
Sbjct 288 VGKNLKEN-----GVDSKRTMIS----VADIRNAFALEKLASVTMRAGKTYKEQMEAHFG 338
Query 370 VDVGENMSGMSTYVGGDSSSLDISEVVN----TNLQSGDVASEAVIA---GKGVGSSQGS 422
+ V E G TY+GG S++ + +V T + D + + GK GS G
Sbjct 339 ISVEEGRDGRCTYIGGFDSNIQVGDVTQSSGTTVTGTKDTSFGGYLGRTTGKATGSGSGH 398
Query 423 EKFEARDWGVLMCIYHNVPLLDYVSSAPDPQFFVTQNTDLPIPELDSIGMQSVPLAMYTN 482
+F+A++ G+LMCIY VP + Y S DP + D +PE +++GMQ PL
Sbjct 399 IRFDAKEHGILMCIYSLVPDVQYDSKRVDPFVQKIERGDFFVPEFENLGMQ--PLFAKNI 456
Query 483 S---DGELVSGFVSPDYTMGYLPRYFSWKTSYDYVLGAFTTTEKEWVAPISSALWKNMLS 539
S + + + G+ PRY +KT+ D G F E ++ A ++M
Sbjct 457 SYKYNNNTANSRIKNLGAFGWQPRYSEYKTALDINHGQFVHQEPLSYWTVARARGESM-- 514
Query 540 TITVRNPQFTYNFFKVNPSVLDSIFQVNADSKWDTDPFLINCAFDVKVVRNLDYSGMP 597
F + FK+NP LD +F VN + TD C F++ V ++ GMP
Sbjct 515 ------SNFNISTFKINPKWLDDVFAVNYNGTELTDQVFGGCYFNIVKVSDMSIDGMP 566
Lambda K H a alpha
0.319 0.133 0.413 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 4446915032268