bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: All non-redundant GenBank CDS translations+PDB+SwissProt+PIR+PRF
excluding environmental samples from WGS projects
49,011,213 sequences; 17,563,301,199 total letters
Query= Contig-51_CDS_annotation_glimmer3.pl_2_1
Length=379
Score E
Sequences producing significant alignments: (Bits) Value
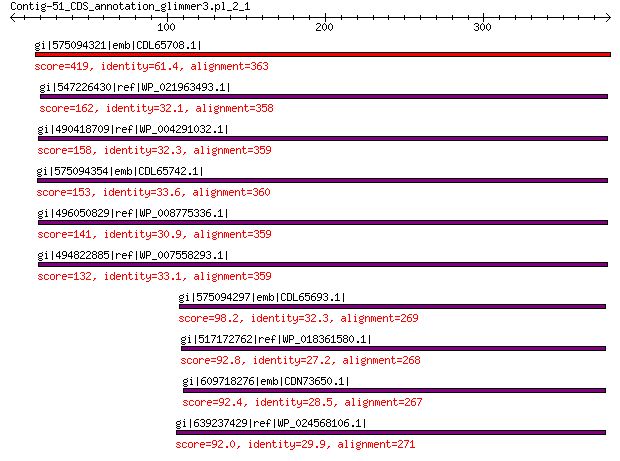
gi|575094321|emb|CDL65708.1| unnamed protein product 419 5e-138
gi|547226430|ref|WP_021963493.1| putative uncharacterized protein 162 3e-41
gi|490418709|ref|WP_004291032.1| hypothetical protein 158 9e-40
gi|575094354|emb|CDL65742.1| unnamed protein product 153 5e-38
gi|496050829|ref|WP_008775336.1| hypothetical protein 141 5e-34
gi|494822885|ref|WP_007558293.1| hypothetical protein 132 1e-30
gi|575094297|emb|CDL65693.1| unnamed protein product 98.2 4e-19
gi|517172762|ref|WP_018361580.1| hypothetical protein 92.8 3e-17
gi|609718276|emb|CDN73650.1| conserved hypothetical protein 92.4 3e-17
gi|639237429|ref|WP_024568106.1| hypothetical protein 92.0 4e-17
>gi|575094321|emb|CDL65708.1| unnamed protein product [uncultured bacterium]
Length=642
Score = 419 bits (1078), Expect = 5e-138, Method: Compositional matrix adjust.
Identities = 223/381 (59%), Positives = 264/381 (69%), Gaps = 24/381 (6%)
Query 17 KSNMLDMRFSNLPLDYFNGVLPTPQFGSESVVSL-------SQNADVYTGFDKSQWQTLD 69
K N+LDMRFSNLPLDYF GVLPT QFGSESVV+L S + T D +W+T
Sbjct 268 KLNLLDMRFSNLPLDYFTGVLPTSQFGSESVVNLNLGNASGSAVLNGTTSKDSGRWRTTT 327
Query 70 Gsafpsgsvsssnsdrsl--tANGKSIEHVHILPSG-----SITSSLSIAALRQATALQK 122
G V+SS + +NG I H H S++ +LSI ALR A A QK
Sbjct 328 GEWEMEQRVASSANGNLKLDNSNGTFISHDHTFSGNVAINTSLSGNLSIIALRNALAAQK 387
Query 123 YKEIQLANDPDFESQIEAHFGIKPKHDMHKSRFIGGSSSMIDINPVVNQNLGAGQNQDNQ 182
YKEIQLAND DF+SQ+EAHFGIKP S FIGGSSSMI+IN +NQNL DN+
Sbjct 388 YKEIQLANDVDFQSQVEAHFGIKPDEKNENSLFIGGSSSMININEQINQNLSG----DNK 443
Query 183 AVTKAAPTGQGGASFKFTADTFGVVIGIYRCTPVLDYSHVGIDRTLLKTDASDFVIPELD 242
A AAP G G AS KFTA T+GVVIGIYRCTPVLD++H+GIDRTL KTDASDFVIPE+D
Sbjct 444 ATYGAAPQGNGSASIKFTAKTYGVVIGIYRCTPVLDFAHLGIDRTLFKTDASDFVIPEMD 503
Query 243 SIGMQQTFQCELFAPT---SQMTA-SAPDKRKYDMSRTFGYAPRYSEYKVSFDRYNGAFC 298
SIGMQQTF+CE+ AP + A D DMS T+GYAPRYSE+K S+DRYNGAFC
Sbjct 504 SIGMQQTFRCEVAAPAPYNDEFKAFRVGDGSSPDMSETYGYAPRYSEFKTSYDRYNGAFC 563
Query 299 DTLKSWVTGFNTHIFDSDRWNDRSYFSISVPQLFVCRPDIVKDIFALQTYHDSNDDNLYV 358
+LKSWVTG N ++ WN ++ I+ P +F CRPDIVK++F + + ++S+DD LYV
Sbjct 564 HSLKSWVTGINFDAIQNNVWN--TWAGINAPNMFACRPDIVKNLFLVSSTNNSDDDQLYV 621
Query 359 GMVNMCYATRNLSRYGLPYSN 379
GMVNMCYATRNLSRYGLPYSN
Sbjct 622 GMVNMCYATRNLSRYGLPYSN 642
>gi|547226430|ref|WP_021963493.1| putative uncharacterized protein [Prevotella sp. CAG:1185]
gi|524103382|emb|CCY83994.1| putative uncharacterized protein [Prevotella sp. CAG:1185]
Length=573
Score = 162 bits (411), Expect = 3e-41, Method: Compositional matrix adjust.
Identities = 115/363 (32%), Positives = 173/363 (48%), Gaps = 32/363 (9%)
Query 20 MLDMRFSNLPLDYFNGVLPTPQFGSESVVSLSQNADVYTGFDKSQWQTLDGsafpsgsvs 79
M D+ + N DYF G+LP Q+G SV S ++ D +L
Sbjct 238 MFDLNYCNFQKDYFTGMLPRAQYGDVSVAS-----PIFGDLDIGDSSSL----------- 281
Query 80 ssnsdrsltANGKSIEHVHILPSGSITSSLSIAALRQATALQKYKEIQLANDPDFESQIE 139
+ S AN + + + + T+ LS+ ALRQA LQK++EI + D+++Q++
Sbjct 282 TFASAPQQGANTIQSGVLVVNNNSNTTAGLSVLALRQAECLQKWREIAQSGKMDYQTQMQ 341
Query 140 AHFGIKPKHDMH-KSRFIGGSSSMIDINPVVNQNLGAGQNQDNQAVTKAAPTGQ-GGASF 197
HF + P + +++GG +S +DI+ VVN NL DNQA + TG G
Sbjct 342 KHFNVSPSATLSGHCKYLGGWTSNLDISEVVNTNLTG----DNQADIQGKGTGTLNGNKV 397
Query 198 KFTADTFGVVIGIYRCTPVLDYSHVGIDRTLLKTDASDFVIPELDSIGMQQTFQCELFAP 257
F + G+++ IY C P+LD+S I R KT +D+ IPE DS+GMQQ + E+
Sbjct 398 DFESSEHGIIMCIYHCLPLLDWSINRIARQNFKTTFTDYAIPEFDSVGMQQLYPSEMIFG 457
Query 258 TSQMTASAPDKRKYDMSRTFGYAPRYSEYKVSFDRYNGAFCDTLKSWVTGFNTHIFDSDR 317
+ + D +M GY PRY++ K S D +G+F DTL SWV+ + R
Sbjct 458 LEDLPS---DPSSINM----GYVPRYADLKTSIDEIHGSFIDTLVSWVSPLTDSYISAYR 510
Query 318 --WNDRSYFSISVP-QLFVCRPDIVKDIFALQTYHDSNDDNLYVGMVNMCYATRNLSRYG 374
D + I++ F P IV +IF ++ N D L + A RN G
Sbjct 511 QACKDAGFSDITMTYNFFKVNPHIVDNIFGVKADSTINTDQLLINSYFDIKAVRNFDYNG 570
Query 375 LPY 377
LPY
Sbjct 571 LPY 573
>gi|490418709|ref|WP_004291032.1| hypothetical protein [Bacteroides eggerthii]
gi|217986636|gb|EEC52970.1| putative capsid protein (F protein) [Bacteroides eggerthii DSM
20697]
Length=578
Score = 158 bits (400), Expect = 9e-40, Method: Compositional matrix adjust.
Identities = 116/371 (31%), Positives = 167/371 (45%), Gaps = 42/371 (11%)
Query 19 NMLDMRFSNLPLDYFNGVLPTPQFGSESVVSLSQNADVYTGFDKSQWQTLDGsafpsgsv 78
N D+R+ N D F+GVLP Q+G +V S++ DV S + T+
Sbjct 238 NFFDLRYCNWQKDLFHGVLPHQQYGETAVASIT--PDVTGKLTLSNFSTVG--------- 286
Query 79 sssnsdrsltANGKSIEHVHILPSGSITSSLSIAALRQATALQKYKEIQLANDPDFESQI 138
TA+G + ++ LP+ LSI LRQA LQK+KEI + + D++ Q+
Sbjct 287 -----TSPTTASGTATKN---LPAFDTVGDLSILVLRQAEFLQKWKEITQSGNKDYKDQL 338
Query 139 EAHFGIKPKHDMHK-SRFIGGSSSMIDINPVVNQNLGAGQNQDNQAVTKAAPTGQGGASF 197
E H+G+ + ++GG SS IDIN V+N N+ D K G +F
Sbjct 339 EKHWGVSVGDGFSELCTYLGGVSSSIDINEVINTNITGSAAAD--IAGKGVGVANGEINF 396
Query 198 KFTADTFGVVIGIYRCTPVLDYSHVGIDRTLLKTDASDFVIPELDSIGMQQTFQCELFAP 257
+ +G+++ IY C P+LDY+ +D LK +++D+ IPE D +GMQ +L P
Sbjct 397 N-SNGRYGLIMCIYHCLPLLDYTTDMLDPAFLKVNSTDYAIPEFDRVGMQSMPLVQLMNP 455
Query 258 TSQMTASAPDKRKYDMSRTFGYAPRYSEYKVSFDRYNGAFCDTLKSWVTGF-NTHIFDSD 316
++ GY PRY +YK S D+ G F TL SWV + N +
Sbjct 456 LRSFANAS--------GLVLGYVPRYIDYKTSVDQSVGGFKRTLNSWVISYGNISVLKQV 507
Query 317 RW-NDRSYFSISVP---------QLFVCRPDIVKDIFALQTYHDSNDDNLYVGMVNMCYA 366
ND S P F PD + IFA+Q D+N D A
Sbjct 508 TLPNDAPPIEPSEPVPSVAPMNFTFFKVNPDCLDPIFAVQAGDDTNTDQFLCSSFFDIKA 567
Query 367 TRNLSRYGLPY 377
RNL GLPY
Sbjct 568 VRNLDTDGLPY 578
>gi|575094354|emb|CDL65742.1| unnamed protein product [uncultured bacterium]
Length=615
Score = 153 bits (387), Expect = 5e-38, Method: Compositional matrix adjust.
Identities = 121/404 (30%), Positives = 182/404 (45%), Gaps = 61/404 (15%)
Query 18 SNMLDMRFSNLPLDYFNGVLPTPQFGSESVVSLSQNADVYTGFDK--------------- 62
+N D+R+ N D F+GVLP Q+GS SVV ++ +V + D
Sbjct 229 NNFFDIRYCNYQKDMFHGVLPVAQYGSASVVPINGQLNVISNGDSGPIFKTSTPDPGTPG 288
Query 63 SQWQTLDGsafpsgsvsssnsdrsltANGKSIEHV-HILPSGSITSSL------------ 109
+ + T+ G+ + GKS + + PS + T SL
Sbjct 289 TSYVTVGGNIGVDNRSFGVSGSTLNV--GKSADPSGYGFPSNASTRSLLWENPNLIIENN 346
Query 110 -----SIAALRQATALQKYKEIQLANDPDFESQIEAHFGIKPKHDM-HKSRFIGGSSSMI 163
I ALRQA LQK+KE+ ++ + D++SQIE H+GIK + H++R++GG ++ +
Sbjct 347 QGFYVPILALRQAEFLQKWKEVSVSGEEDYKSQIEKHWGIKVSDFLSHQARYLGGCATSL 406
Query 164 DINPVVNQNLGAGQNQDNQAVTKAAPTGQGGASFKFTAD-TFGVVIGIYRCTPVLDYSHV 222
DIN V+N N+ DN A T G S +F + +G+++ IY P++DY
Sbjct 407 DINEVINNNITG----DNAADIAGKGTFTGNGSIRFESKGEYGIIMCIYHVLPIVDYVGS 462
Query 223 GIDRTLLKTDASDFVIPELDSIGMQQTFQCELFAPTSQMTASAPDKRKYDMSRTF-GYAP 281
G+D + DA+ F IPELD IGM+ P + + D TF GYAP
Sbjct 463 GVDHSCTLVDATSFPIPELDQIGMESVPLVRAMNPVKESDTPSAD--------TFLGYAP 514
Query 282 RYSEYKVSFDRYNGAFCDTLKSW--------VTGFNTHIFDSDRWNDRSYFSISVPQLFV 333
RY ++K S DR G F D+L++W +T N+ F S+ + + F
Sbjct 515 RYIDWKTSVDRSVGDFADSLRTWCLPVGDKELTSANSLNFPSNPNVEPDSIAAG---FFK 571
Query 334 CRPDIVKDIFALQTYHDSNDDNLYVGMVNMCYATRNLSRYGLPY 377
P IV +FA+ D RNL GLPY
Sbjct 572 VNPSIVDPLFAVVADSTVKTDEFLCSSFFDVKVVRNLDVNGLPY 615
>gi|496050829|ref|WP_008775336.1| hypothetical protein [Bacteroides sp. 2_2_4]
gi|229448893|gb|EEO54684.1| putative capsid protein (F protein) [Bacteroides sp. 2_2_4]
Length=580
Score = 141 bits (356), Expect = 5e-34, Method: Compositional matrix adjust.
Identities = 111/365 (30%), Positives = 173/365 (47%), Gaps = 36/365 (10%)
Query 19 NMLDMRFSNLPLDYFNGVLPTPQFGSESVVSLSQNADVYTGFDKSQWQTLDGsafpsgsv 78
NM D+R+ N D F+GVLP Q+G + V+++ ++V + + QT DG
Sbjct 246 NMFDLRYCNWQKDLFHGVLPRQQYGDTAAVNVNL-SNVLSA--QYMVQTPDGDPVGGSPF 302
Query 79 sssnsdrsltANGKSIEHVHILPSGSITSSLSIAALRQATALQKYKEIQLANDPDFESQI 138
S + G +++ V+ SG+ T + ALRQA LQK+KEI + + D++ QI
Sbjct 303 S---------STGVNLQTVN--GSGTFT----VLALRQAEFLQKWKEITQSGNKDYKDQI 347
Query 139 EAHFGIKPKHDMHK-SRFIGGSSSMIDINPVVNQNLGAGQNQDNQAVTKAAPTGQGGASF 197
E H+ + + S ++GG+++ +DIN VVN N+ D K G G SF
Sbjct 348 EKHWNVSVGEAYSEMSLYLGGTTASLDINEVVNNNITGSNAAD--IAGKGVVVGNGRISF 405
Query 198 KFTADTFGVVIGIYRCTPVLDYSHVGIDRTLLKTDASDFVIPELDSIGMQQTFQCELFAP 257
+ +G+++ IY P+LDY+ ++ K +++DF IPE D +GM+ L P
Sbjct 406 D-AGERYGLIMCIYHSLPLLDYTTDLVNPAFTKINSTDFAIPEFDRVGMESVPLVSLMNP 464
Query 258 TSQMTASAPDKRKYDM-SRTFGYAPRYSEYKVSFDRYNGAFCDTLKSWVTGF-NTHIFDS 315
+ Y++ S GYAPRY YK D GAF TLKSWV + N + +
Sbjct 465 L---------QSSYNVGSSILGYAPRYISYKTDVDSSVGAFKTTLKSWVMSYDNQSVINQ 515
Query 316 DRWND---RSYFSISVPQLFVCRPDIVKDIFALQTYHDSNDDNLYVGMVNMCYATRNLSR 372
+ D S ++ F P+ V +FA+ + + D RNL
Sbjct 516 LNYQDDPNNSPGTLVNYTNFKVNPNCVDPLFAVAASNSIDTDQFLCSSFFDVKVVRNLDT 575
Query 373 YGLPY 377
GLPY
Sbjct 576 DGLPY 580
>gi|494822885|ref|WP_007558293.1| hypothetical protein [Bacteroides plebeius]
gi|198272099|gb|EDY96368.1| putative capsid protein (F protein) [Bacteroides plebeius DSM
17135]
Length=613
Score = 132 bits (332), Expect = 1e-30, Method: Compositional matrix adjust.
Identities = 119/380 (31%), Positives = 176/380 (46%), Gaps = 33/380 (9%)
Query 19 NMLDMRFSNLPLDYFNGVLPTPQFGSESVVSLSQNADVYTGFDKSQWQT-LDGsafpsgs 77
N+ DMR+SN D +G +P Q+G S V +S + V G + T DG AF +G+
Sbjct 246 NLFDMRYSNWQRDLLHGTIPQAQYGEASAVPVSGSMQVVEGPTPPAFTTGQDGVAFLNGN 305
Query 78 vsssnsdrsltANGKSIEHVHILPSGSITSSL----------SIAALRQATALQKYKEIQ 127
V+ S L A S+ IL + S L SI ALR+A A QK+KE+
Sbjct 306 VTIQGSSGYLQAQ-TSVGESRILRFNNTNSGLIVEGDSSFGVSILALRRAEAAQKWKEVA 364
Query 128 LANDPDFESQIEAHFGI---KPKHDMHKSRFIGGSSSMIDINPVVNQNLGAGQNQDNQAV 184
LA++ D+ SQIEAH+G K DM +++G + + IN VVN N+ G+N + A
Sbjct 365 LASEEDYPSQIEAHWGQSVNKAYSDM--CQWLGSINIDLSINEVVNNNI-TGENAADIA- 420
Query 185 TKAAPTGQGGASFKFTADTFGVVIGIYRCTPVLDYSHVGIDRTLLKTDASDFVIPELDSI 244
K +G G +F +G+V+ ++ P LDY T+ DF IPE D I
Sbjct 421 GKGTMSGNGSINFN-VGGQYGIVMCVFHVLPQLDYITSAPHFGTTLTNVLDFPIPEFDKI 479
Query 245 GMQQTFQCELFAPTSQMTASAPDKRKYDMSRT--FGYAPRYSEYKVSFDRYNGAFCDTLK 302
GM+Q P P + +S FGYAP+Y +K + D+ G F +LK
Sbjct 480 GMEQVPVIRGLNPVK------PKDGDFKVSPNLYFGYAPQYYNWKTTLDKSMGEFRRSLK 533
Query 303 SWVTGFNTHIF---DSDRWNDRSYFSISVPQ--LFVCRPDIVKDIFALQTYHDSNDDNLY 357
+W+ F+ DS + D + F P ++ ++FA++ D N D
Sbjct 534 TWIIPFDDEALLAADSVDFPDNPNVEADSVKAGFFKVSPSVLDNLFAVKANSDLNTDQFL 593
Query 358 VGMVNMCYATRNLSRYGLPY 377
+ R+L GLPY
Sbjct 594 CSTLFDVNVVRSLDPNGLPY 613
>gi|575094297|emb|CDL65693.1| unnamed protein product [uncultured bacterium]
Length=630
Score = 98.2 bits (243), Expect = 4e-19, Method: Compositional matrix adjust.
Identities = 87/301 (29%), Positives = 126/301 (42%), Gaps = 47/301 (16%)
Query 108 SLSIAALRQATALQKYKEIQLANDPDFESQIEAHFGIK-PKHDMHKSRFIGGSSSMIDIN 166
+L+++ LR A K I +++Q AHFG K P+ + ++GG S + I+
Sbjct 343 TLNVSQLRALYATDKLLRITQFAGKHYDAQTLAHFGKKVPQGVSGEVYYLGGQSQRLQIS 402
Query 167 PVVNQNLGAGQNQDN------QAVTKAAPTGQGGASFKFTADTFGVVIGIYRCTPVLDYS 220
P+ L +GQ D + +AA QG F F A G+++ IY P +YS
Sbjct 403 PITA--LSSGQTSDGSDTVFGEQGARAASVTQGQKPFTFEAPCHGILMAIYSAVPEANYS 460
Query 221 HVGIDRTLLKTDASDFVIPELDSIGMQQTFQCELFAPTSQMTASAPDKRKY-DMSRTFGY 279
IDR ++DF PELD+IGM + E P + + P D +++ G+
Sbjct 461 CDAIDRINTLAYSNDFYKPELDNIGMSPLYSYEFSVPGYTLFRNPPTPYSSDDAAQSLGW 520
Query 280 APRYSEYKVSFDRYNGAFCDTLKSWVTGFNTHIFDSDRWNDRSYFSI---SVPQLF---- 332
RYS +K DR GA TL+SW R Y ++ S PQLF
Sbjct 521 QFRYSWFKTKVDRTCGALNRTLRSWCP-------------KRDYLALGLQSRPQLFNYAS 567
Query 333 --VCRPDIVKDIFALQ-------TYHDSND--------DNLYVGMVNMCYATRNLSRYGL 375
P + +F L Y +D D L M CY T +S YGL
Sbjct 568 LYYVSPSYLDGLFYLNFAPPIDYRYPVDSDMSSIAFETDPLIHDMQITCYKTSVMSTYGL 627
Query 376 P 376
P
Sbjct 628 P 628
>gi|517172762|ref|WP_018361580.1| hypothetical protein [Prevotella nanceiensis]
Length=568
Score = 92.8 bits (229), Expect = 3e-17, Method: Compositional matrix adjust.
Identities = 73/277 (26%), Positives = 118/277 (43%), Gaps = 23/277 (8%)
Query 109 LSIAALRQATALQKYKEIQLANDPDFESQIEAHFGIKPKHDMH-KSRFIGGSSSMIDINP 167
+S+A +R A AL+K + + ++ Q+EAHFGI + + +IGG S I +
Sbjct 304 ISVADIRNAFALEKLASVTMRAGKTYKEQMEAHFGISVEEGRDGRCTYIGGFDSNIQVGD 363
Query 168 VVNQNLGAGQNQDNQAV------TKAAPTGQGGASFKFTADTFGVVIGIYRCTPVLDYSH 221
V + + + T TG G +F A G+++ IY P + Y
Sbjct 364 VTQSSGTTVTGTKDTSFGGYLGRTTGKATGSGSGHIRFDAKEHGILMCIYSLVPDVQYDS 423
Query 222 VGIDRTLLKTDASDFVIPELDSIGMQQTFQCELFAPTSQMTASAPDKRKYDMSRTFGYAP 281
+D + K + DF +PE +++GMQ F + + TA++ K FG+ P
Sbjct 424 KRVDPFVQKIERGDFFVPEFENLGMQPLFAKNISYKYNNNTANSRIKN----LGAFGWQP 479
Query 282 RYSEYKVSFDRYNGAFC--DTLKSWVTGFNTHIFDSDRWNDRSYFSISVPQLFVCRPDIV 339
RYSEYK + D +G F + L W R S F+IS F P +
Sbjct 480 RYSEYKTALDINHGQFVHQEPLSYWTVA-------RARGESMSNFNIST---FKINPKWL 529
Query 340 KDIFALQTYHDSNDDNLYVGMVNMCYATRNLSRYGLP 376
D+FA+ D ++ G ++S G+P
Sbjct 530 DDVFAVNYNGTELTDQVFGGCYFNIVKVSDMSIDGMP 566
>gi|609718276|emb|CDN73650.1| conserved hypothetical protein [Elizabethkingia anophelis]
Length=537
Score = 92.4 bits (228), Expect = 3e-17, Method: Compositional matrix adjust.
Identities = 76/269 (28%), Positives = 115/269 (43%), Gaps = 27/269 (10%)
Query 110 SIAALRQATALQKYKEIQLANDPDFESQIEAHFGIKPKHD-MHKSRFIGGSSSMIDINPV 168
++ LR+A LQ++ E + I + FG+K + + F+GG+ S I I+ V
Sbjct 290 TVNDLRRAFKLQEWLEKNARAGSRYAESILSFFGVKTSDGRLQRPEFLGGNKSPIMISEV 349
Query 169 VNQNLGAGQNQDNQAVTKAAPTGQGGASFKFTADTFGVVIGIYRCTPVLDYSHVGIDRTL 228
+ Q+ G+ G +F + G VIG+ P YS GI R
Sbjct 350 LQQSATDSTTPQGNMAGHGIGIGKDGGFSRFFEEH-GYVIGLMSVIPKTSYSQ-GIPRHF 407
Query 229 LKTDASDFVIPELDSIGMQQTFQCELFAPTSQMTASAPDKRKYDMSRTFGYAPRYSEYKV 288
K+D D+ P+ + IG Q + E+FA +D FGY PRYSEYK
Sbjct 408 SKSDKFDYFWPQFEHIGEQPVYNKEIFAKNID---------AFDSEAVFGYLPRYSEYKF 458
Query 289 SFDRYNGAFCDTLKSWVTGFNTHIFDSDRWNDRSYFSISVPQLFV-CRPDIVKDIFALQT 347
S +G F D L W G IFD+D+ + Q F+ C + + IFA++
Sbjct 459 SPSTVHGDFKDDLYFWHLG---RIFDTDK-------PPVLNQSFIECDKNALSRIFAVE- 507
Query 348 YHDSNDDNLYVGMVNMCYATRNLSRYGLP 376
+ D Y + A R +S +G P
Sbjct 508 ---DDTDKFYCHLYQKITAKRKMSYFGDP 533
>gi|639237429|ref|WP_024568106.1| hypothetical protein [Elizabethkingia anophelis]
Length=546
Score = 92.0 bits (227), Expect = 4e-17, Method: Compositional matrix adjust.
Identities = 81/272 (30%), Positives = 113/272 (42%), Gaps = 25/272 (9%)
Query 106 TSSLSIAALRQATALQKYKEIQLANDPDFESQIEAHFGIKPKHD-MHKSRFIGGSSSMID 164
S +I LR+A LQ++ E + I + FG+K + + F+GG+ + I
Sbjct 295 ASGSTINDLRRAFKLQEWLEKNARAGSRYAESILSFFGVKTSDGRLQRPEFLGGNKTPIL 354
Query 165 INPVVNQNLGAGQNQDNQAVTKAAPTGQGGASFKFTADTFGVVIGIYRCTPVLDYSHVGI 224
I+ V+ Q+ G+ G KF + G VIG+ P YS GI
Sbjct 355 ISEVLQQSSTDSTTPQGNMAGHGISVGKEGGFSKFFEEH-GYVIGLMSVIPKTSYSQ-GI 412
Query 225 DRTLLKTDASDFVIPELDSIGMQQTFQCELFAPTSQMTASAPDKRKYDMSRTFGYAPRYS 284
R K D D+ P+ + IG Q + E+FA YD FGY PRYS
Sbjct 413 PRHFSKFDKFDYFWPQFEHIGEQPVYNKEIFAKNVG---------DYDSGGVFGYVPRYS 463
Query 285 EYKVSFDRYNGAFCDTLKSWVTGFNTHIFDSDRWNDRSYFSISVPQLFVCRPDIVKDIFA 344
EYK S +G F DTL W G IFDS + I V + + R IFA
Sbjct 464 EYKYSPSTIHGDFKDTLYFWHLG---RIFDSSAPPKLNRDFIEVNKSGLSR------IFA 514
Query 345 LQTYHDSNDDNLYVGMVNMCYATRNLSRYGLP 376
++ N D Y + A R +S +G P
Sbjct 515 VE----DNSDKFYCHLYQKITAKRKMSYFGDP 542
Lambda K H a alpha
0.318 0.133 0.397 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 2359945006260