bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: All non-redundant GenBank CDS translations+PDB+SwissProt+PIR+PRF
excluding environmental samples from WGS projects
49,011,213 sequences; 17,563,301,199 total letters
Query= Contig-5_CDS_annotation_glimmer3.pl_2_3
Length=368
Score E
Sequences producing significant alignments: (Bits) Value
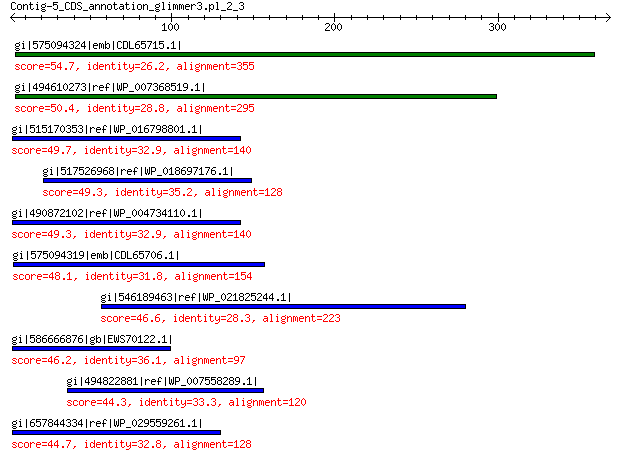
gi|575094324|emb|CDL65715.1| unnamed protein product 54.7 4e-05
gi|494610273|ref|WP_007368519.1| hypothetical protein 50.4 0.001
gi|515170353|ref|WP_016798801.1| hypothetical protein 49.7 0.002
gi|517526968|ref|WP_018697176.1| hypothetical protein 49.3 0.003
gi|490872102|ref|WP_004734110.1| hypothetical protein 49.3 0.003
gi|575094319|emb|CDL65706.1| unnamed protein product 48.1 0.005
gi|546189463|ref|WP_021825244.1| hypothetical protein 46.6 0.015
gi|586666876|gb|EWS70122.1| hypothetical protein Y702_04245 46.2 0.023
gi|494822881|ref|WP_007558289.1| hypothetical protein 44.3 0.082
gi|657844334|ref|WP_029559261.1| hypothetical protein 44.7 0.083
>gi|575094324|emb|CDL65715.1| unnamed protein product [uncultured bacterium]
Length=370
Score = 54.7 bits (130), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 93/377 (25%), Positives = 153/377 (41%), Gaps = 52/377 (14%)
Query 4 LIGAAIGAISQNFNTSESIAANKQEQQSNRDYNLNLARLQNQWNREQWEREAQYNSPAAY 63
L+G A I +I A ++E + R++ +A Q RE E YN+P+A
Sbjct 13 LLGGAFSGIGAKKRQEAAIKAQREENEKARNWQQKMAEWQVGIERENLADERAYNNPSAV 72
Query 64 RARLASAGMNSDLAY-SNVNGVAPAS-PGMTSGEPSSPVDYSA-IAGKQTIGSAVSQALA 120
RL AG+N DL Y S +G+ ++ G S P D + I G T+ ++ Q A
Sbjct 73 MKRLKDAGLNPDLMYGSGASGLVDSNVAGSASVGNVPPADVAGPIMGTPTMMESLFQGAA 132
Query 121 NEQARANIALTQAQKNKTDEEAGKTSEEAEGVRIDNLTRGASNTLEIQLKEGVIKLNDSV 180
A T A+ + K E + IDN + AS+ I+L ++L +
Sbjct 133 -------YAKTVAETKNIKADTSKKEGEVTSLNIDNFVKAASSDNAIKLSGVEVQLTKAQ 185
Query 181 kqlneqnkknlqqllenlklesnnlAEQWQVIRETW---SNLRVDRAL---------KMI 228
+ + K L + ++ N L Q I ETW SNL L +
Sbjct 186 AEYTSEQKTKLISEINDINEHVNLLKAQ---ISETWARTSNLDSSTVLNRTTAILNNRRF 242
Query 229 DLKFREKQNVAILKKISSETNLNYVQASSM-------TKRLMLDMALGKTQMNLMTQQAI 281
DL+ E +++ ++ NL+ +A S+ + D AL + + L
Sbjct 243 DLECEEFARR--VRETDAKVNLSEAEAKSILVTMYAKVNNIDADTALKQANIRL------ 294
Query 282 TETQKRVNMRTEDWYNRGRIRSVYLQNGQLSFDLSQSIKWDDTMKSIDWCEKFCRSIFML 341
T+ QK + E + N S+ + F L Q K+DD + + + +S++ +
Sbjct 295 TDAQK---TQVEHYTN-----SIDIHRDAAVFKLQQDQKYDDAQRIVSVANQATQSLYHI 346
Query 342 TQSFQGAASGSLPKTGG 358
+Q AS LP GG
Sbjct 347 SQ----VASDWLPSPGG 359
>gi|494610273|ref|WP_007368519.1| hypothetical protein [Prevotella multiformis]
gi|324988545|gb|EGC20508.1| hypothetical protein HMPREF9141_0987 [Prevotella multiformis
DSM 16608]
Length=437
Score = 50.4 bits (119), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 85/313 (27%), Positives = 135/313 (43%), Gaps = 47/313 (15%)
Query 4 LIGAAIGAISQNFNTSESIAANKQEQQSNRDYNLNLARL---QNQWNREQWEREAQYNSP 60
L+G G S N + AN+ Q R+ N N ++ QN +N W + QYNSP
Sbjct 79 LLGGLFGGHS---NKTAQDRANETNLQIAREANQNQYQMFQEQNAFNERMWNQMNQYNSP 135
Query 61 AAYRARLASAGMNSDLAYSNVNGVAPASPGMTSGEPSSPVDYSAIAGKQTIGSAVSQALA 120
AA R AG+N +A NV S ++ P V + + +G AV + A
Sbjct 136 AAQMQRYTDAGINPYIAAGNVQTGNAQSALQSAPAPQQHV--AQVMPATGMGDAVQNSFA 193
Query 121 ------NEQARANIALTQAQKNKTDEEAGKTSEEAEGVRIDNLTRG--ASNTLEIQLKEG 172
++ A+ +AL QA+ KTD EA R+++ G + TL I +
Sbjct 194 QIGNVISQFAQNQLALAQAK--KTDAEASWID------RLNSAQMGKLGAETLNIHNQNS 245
Query 173 VIKLNDSVkqlne--qnkknlqqllenlklesnnlAEQWQVIRETWSNLRVDRAL--KMI 228
++ L+ +K + + + + A+ + + E+ D A+ I
Sbjct 246 LLGLDYQIKSDTLGNYKLLSDLSVQQAALTNNLVDAQTRKALFES------DLAMVESHI 299
Query 229 DLKFREKQNVAILKKIS---SETNLNYVQASSMTKRLMLDMALGKTQMNLMTQQAITETQ 285
K+ EKQ +L++IS S LNYV A + K L Q NLM +QA TE +
Sbjct 300 KAKYGEKQ---VLQEISESVSRQYLNYVSAHNSEK-------LTTAQCNLMFEQAKTEVE 349
Query 286 KRVNMRTEDWYNR 298
KR +R ++ +R
Sbjct 350 KRFGIRLDNDTSR 362
>gi|515170353|ref|WP_016798801.1| hypothetical protein [Vibrio cyclitrophicus]
Length=395
Score = 49.7 bits (117), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 46/154 (30%), Positives = 72/154 (47%), Gaps = 25/154 (16%)
Query 2 YGLIGAAIGAISQNFNTSESIAANKQEQQSNRDYNLNLARLQNQWNREQWEREAQYNSPA 61
Y ++ AI I+ N +E + +++SN ++ L + R QW A+++ A
Sbjct 175 YHILPTAISGIAAAMNPAEWLTEAGSDKESNDEFRLRI--------RNQWSAVAKWHIDA 226
Query 62 AYRARLAS-AGMNSDLAYSNVNGVAPASPG-------MTSGEPSSPV-----DYSAIAGK 108
AYRA L S AG+N D Y N AP PG + +GEPSS + +Y G+
Sbjct 227 AYRALLTSRAGINDDNVYFQHN--APRGPGTANAYILLDTGEPSSEMLADLNEYIRSQGQ 284
Query 109 QTIGSAVSQALANEQARANIAL-TQAQKNKTDEE 141
G + Q +A + AN+ Q++ T EE
Sbjct 285 HGHGDDL-QVMAMAETEANVVCRIWPQRSLTMEE 317
>gi|517526968|ref|WP_018697176.1| hypothetical protein [Alistipes onderdonkii]
Length=364
Score = 49.3 bits (116), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 45/143 (31%), Positives = 64/143 (45%), Gaps = 28/143 (20%)
Query 21 SIAANKQEQQSNRDYNLNLARLQNQWNREQWEREAQYNSPAAYRARLASAGMNSDLAYSN 80
S+A NK+++ ++Y+ L+ L+ + N+E W + YNSP+A R AGM+ S
Sbjct 37 SVAENKRQRAFQKEYSKYLSELEAKQNQEYWNK---YNSPSAQRIARMKAGMSPFADESG 93
Query 81 VNGVA--------------PAS-PGMTSGEPSSPVDYSAIAGKQTIGSAVSQALANEQAR 125
V + P + PG + P SP S V Q L+ QA
Sbjct 94 VQAMGVDPGSYSGSSPSSQPFTQPGGNAMSPLSPA----------FASGVQQVLSARQAE 143
Query 126 ANIALTQAQKNKTDEEAGKTSEE 148
ANI LT A KT EA K +E
Sbjct 144 ANIQLTDANTAKTQAEAVKAQQE 166
>gi|490872102|ref|WP_004734110.1| hypothetical protein [Vibrio splendidus]
gi|84376029|gb|EAP92917.1| hypothetical protein V12B01_20762 [Vibrio splendidus 12B01]
Length=395
Score = 49.3 bits (116), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 46/154 (30%), Positives = 72/154 (47%), Gaps = 25/154 (16%)
Query 2 YGLIGAAIGAISQNFNTSESIAANKQEQQSNRDYNLNLARLQNQWNREQWEREAQYNSPA 61
Y ++ AI I+ N +E + +++SN ++ L + R QW A+++ A
Sbjct 175 YHILPTAISGIAAAMNPAEWLTEAGADKESNDEFRLRI--------RNQWSAVAKWHIDA 226
Query 62 AYRARLAS-AGMNSDLAYSNVNGVAPASPG-------MTSGEPSSPV-----DYSAIAGK 108
AYRA L S AG+N D Y N AP PG + +GEPSS + +Y G+
Sbjct 227 AYRALLTSRAGINDDNVYFQHN--APRGPGTANAYILLDTGEPSSEMLADLNEYIRSQGQ 284
Query 109 QTIGSAVSQALANEQARANIAL-TQAQKNKTDEE 141
G + Q +A + AN+ Q++ T EE
Sbjct 285 HGHGDDL-QVMALPETEANVVCRIWPQRSLTMEE 317
>gi|575094319|emb|CDL65706.1| unnamed protein product [uncultured bacterium]
Length=396
Score = 48.1 bits (113), Expect = 0.005, Method: Compositional matrix adjust.
Identities = 49/160 (31%), Positives = 77/160 (48%), Gaps = 12/160 (8%)
Query 3 GLIGAAIGAISQNFNTSESIAANKQEQQSNRDYN---LNLARLQNQWNREQWEREAQYNS 59
GLI A I+ F+++ S A K + Q+ R+ N +A N++N W + +YN
Sbjct 27 GLIAGAGSLINGLFSSNGSKQAAKYQLQAVRETNQANREIADQNNKFNERMWNLQNEYNR 86
Query 60 PAAYRARLASAGMNSDLAYSNVN-GVAPASPGM-TSGEPSSPVDYSAIAGK-QTIGSAVS 116
P RARL +AG+N L + G+A ++P TSG +P + IAG Q +G+++S
Sbjct 87 PDMQRARLEAAGLNPYLMMDGGSAGIAESAPTADTSGTQIAPDIGNTIAGGYQAMGNSIS 146
Query 117 QALANEQARANIALTQAQKNKTDEEAGKTSEEAEGVRIDN 156
A + IA Q + KT EA+ + N
Sbjct 147 SAA------SQIAQMTFQDDLQKANVAKTVAEAKNAHLQN 180
>gi|546189463|ref|WP_021825244.1| hypothetical protein [Prevotella salivae]
gi|544001992|gb|ERK01416.1| hypothetical protein HMPREF9145_2738 [Prevotella salivae F0493]
Length=322
Score = 46.6 bits (109), Expect = 0.015, Method: Compositional matrix adjust.
Identities = 63/236 (27%), Positives = 106/236 (45%), Gaps = 29/236 (12%)
Query 57 YNSPAAYRARLASAGMNSDLAYSNVNGVAPASPGMTSGEPSSPVDYSAIAGKQTIGSAVS 116
YN+P R +AG+N + VN + TS +SPVDYS+ G Q +G+A +
Sbjct 50 YNTPGMQMQRFQNAGLNPYMMLGQVN-----AGNQTSVASTSPVDYSS--GIQGVGNAAN 102
Query 117 QALANEQARANIALTQAQKNKTDEEAGKTSEEAEGVRIDNLTRGASNTLEIQ--LKEGVI 174
+ A + I + +A KT+ E +ID TR A N I +K+GV+
Sbjct 103 TLIQAASANSQIEVNKATIAKTESETALN-------QIDAQTRAAENAARINQLVKQGVL 155
Query 175 --KLNDSVkqlneqnkknlqqllenlklesnnlAEQWQVIRETWSNLRVDRALKMIDL-- 230
+ D++ Q + +L + L + AEQ + ++ + ++ K +DL
Sbjct 156 TQQQADNLSQQFDLTMLTWDELRKQPGLANLLSAEQIENLKAATGKIGSEK--KGLDLQN 213
Query 231 ----KFREKQNVAILKKISSETNLNYVQASSMTKRLMLD---MALGKTQMNLMTQQ 279
KF EKQ A L S+T +N+ M ++ L+ M L + Q+ ++ Q
Sbjct 214 GITAKFGEKQAAATLGNTLSQTGVNHANIGLMGSQIGLNGTYMDLNRGQLGVLRNQ 269
>gi|586666876|gb|EWS70122.1| hypothetical protein Y702_04245 [Vibrio vulnificus BAA87]
gi|674452857|gb|KFK68383.1| phage protein [Vibrio vulnificus]
Length=395
Score = 46.2 bits (108), Expect = 0.023, Method: Compositional matrix adjust.
Identities = 35/105 (33%), Positives = 52/105 (50%), Gaps = 18/105 (17%)
Query 2 YGLIGAAIGAISQNFNTSESIAANKQEQQSNRDYNLNLARLQNQWNREQWEREAQYNSPA 61
Y ++ AI I+ N +E + A +++SN + L + R QW A+++ A
Sbjct 175 YHILPTAISGIAAATNPAEWLLAAGADKESNDELRLRI--------RNQWSAVARWHIDA 226
Query 62 AYRARLAS-AGMNSDLAYSNVNGVAPASPG-------MTSGEPSS 98
AYRA L S AG+N D Y N AP PG + +GEPS+
Sbjct 227 AYRALLTSRAGINDDNVYFEHN--APRGPGTANAYILLDTGEPST 269
>gi|494822881|ref|WP_007558289.1| hypothetical protein [Bacteroides plebeius]
gi|198272097|gb|EDY96366.1| hypothetical protein BACPLE_00802 [Bacteroides plebeius DSM 17135]
Length=344
Score = 44.3 bits (103), Expect = 0.082, Method: Compositional matrix adjust.
Identities = 40/144 (28%), Positives = 63/144 (44%), Gaps = 26/144 (18%)
Query 36 NLNLARLQNQWNREQWER-----------EAQYNSPAAYRARLASAGMNSDLAYSNVNGV 84
N+ +A++ N++NREQ ER E +YNS ++ R RL AG+N + G
Sbjct 47 NIQIAQMSNEYNREQLERQIEQEWDMWNAENEYNSASSQRKRLEEAGLNPYMMMD--GGS 104
Query 85 APASPGMTS------------GEPSSPVDYSAIAGKQTIGSAVSQAL-ANEQARANIALT 131
A ++ MTS G P D S ++G + I S L A E R +
Sbjct 105 AGSASSMTSPAAQPAVVPQMQGATMQPADMSGLSGLRGIASEFIATLKAQEDIRGQQLIN 164
Query 132 QAQKNKTDEEAGKTSEEAEGVRID 155
+ Q+ + +A K + E R +
Sbjct 165 EGQEIENQYKADKLLADLEKTRTE 188
>gi|657844334|ref|WP_029559261.1| hypothetical protein [Vibrio parahaemolyticus]
Length=395
Score = 44.7 bits (104), Expect = 0.083, Method: Compositional matrix adjust.
Identities = 42/141 (30%), Positives = 65/141 (46%), Gaps = 24/141 (17%)
Query 2 YGLIGAAIGAISQNFNTSESIAANKQEQQSNRDYNLNLARLQNQWNREQWEREAQYNSPA 61
Y ++ AI I N +E + +++SN + L + R QW A+++ A
Sbjct 175 YHILPTAIPGIGAVTNPAEWLNEAGSDKESNDELRLRV--------RNQWSAVARWHIDA 226
Query 62 AYRARLAS-AGMNSDLAYSNVNGVAPASPG-------MTSGEPSSPV-----DYSAIAGK 108
AYR+ L S AG+N D Y N AP PG + +GEPSS + +Y I G+
Sbjct 227 AYRSLLTSRAGINDDNVYFEHN--APRGPGTANALILLDTGEPSSDMLADLNEYIRIEGQ 284
Query 109 QTIGSAVSQALANEQARANIA 129
G + Q LA + +I+
Sbjct 285 HGHGDDL-QVLAMPETTHDIS 304
Lambda K H a alpha
0.312 0.125 0.352 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 2247078071178