bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: All non-redundant GenBank CDS translations+PDB+SwissProt+PIR+PRF
excluding environmental samples from WGS projects
49,011,213 sequences; 17,563,301,199 total letters
Query= Contig-7_CDS_annotation_glimmer3.pl_2_1
Length=277
Score E
Sequences producing significant alignments: (Bits) Value
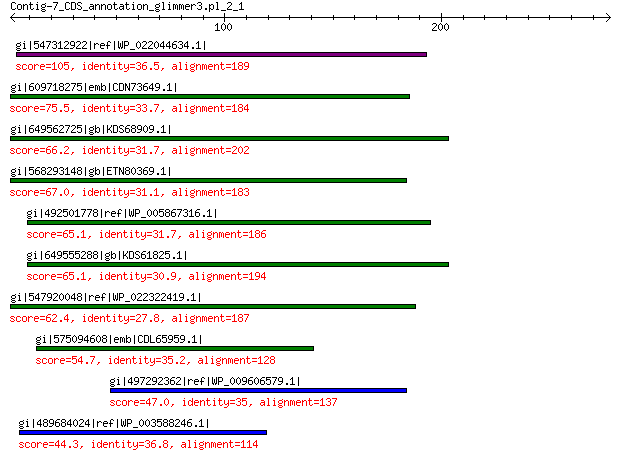
gi|547312922|ref|WP_022044634.1| putative replication initiation... 105 8e-23
gi|609718275|emb|CDN73649.1| conserved hypothetical protein 75.5 6e-13
gi|649562725|gb|KDS68909.1| hypothetical protein M096_3339 66.2 1e-09
gi|568293148|gb|ETN80369.1| hypothetical protein NECAME_18023 67.0 2e-09
gi|492501778|ref|WP_005867316.1| hypothetical protein 65.1 5e-09
gi|649555288|gb|KDS61825.1| hypothetical protein M095_3808 65.1 5e-09
gi|547920048|ref|WP_022322419.1| putative replication protein 62.4 4e-08
gi|575094608|emb|CDL65959.1| unnamed protein product 54.7 2e-05
gi|497292362|ref|WP_009606579.1| hypothetical protein 47.0 0.007
gi|489684024|ref|WP_003588246.1| MULTISPECIES: hypothetical protein 44.3 0.050
>gi|547312922|ref|WP_022044634.1| putative replication initiation protein [Alistipes finegoldii
CAG:68]
gi|524208442|emb|CCZ76638.1| putative replication initiation protein [Alistipes finegoldii
CAG:68]
Length=320
Score = 105 bits (261), Expect = 8e-23, Method: Compositional matrix adjust.
Identities = 69/205 (34%), Positives = 113/205 (55%), Gaps = 30/205 (15%)
Query 4 ELRTEP--NAYFMTLTISDENYEILKNTCKSEDKNTIATKAIRLTLERIRKKTGKSIKHW 61
ELR P F+TLT +D++ E S+D N KA+RL L+R RK GK I+HW
Sbjct 65 ELRKYPPGTCLFVTLTFNDDSLEKF-----SKDTN----KAVRLFLDRFRKVYGKQIRHW 115
Query 62 FITELGHEKTERLHLHGIVWGI-------------GTDQLIKEKWNYGITYTGNFVNEKT 108
F+ E G R H HGI++ + G L+ W YG + G +V+++T
Sbjct 116 FVCEFG-TLHGRPHYHGILFNVPQALIDGYDSDMPGHHPLLASCWKYGFVFVG-YVSDET 173
Query 109 INYITKYMTK-IDEDHPEFVGKVLCSKGIGAGYIKRADASKHKYEKGKTIETYRLRNGAK 167
+YITKY+TK I+ D + +V+ S GIG+ Y+ ++S HK + + + + NG +
Sbjct 174 CSYITKYVTKSINGD--KVRPRVISSFGIGSNYLNTEESSLHKLGNQR-YQPFMVLNGFQ 230
Query 168 INLPIYYRNKLFTEEERELLFIDKI 192
+P YY NK+F++ +++ + +D++
Sbjct 231 QAMPRYYYNKIFSDVDKQNMVVDRL 255
>gi|609718275|emb|CDN73649.1| conserved hypothetical protein [Elizabethkingia anophelis]
Length=265
Score = 75.5 bits (184), Expect = 6e-13, Method: Compositional matrix adjust.
Identities = 62/191 (32%), Positives = 92/191 (48%), Gaps = 17/191 (9%)
Query 1 MSEELRTEPNAYFMTLTISDENYEILKNTCKSEDKNTIATKAIRLTLERIRKKTGKSIKH 60
++EEL+ +A+F+TLT SD N S D + +L ++R RK IK+
Sbjct 43 LTEELKVSKSAHFVTLTYSDVYLPYSDNGLISLD-----YRDFQLFMKRARKLQKSKIKY 97
Query 61 WFITELGHEKTERLHLHGIVWGIGTDQLIKEKWNYGITYTGNFVNEKTINYITKYMTK-I 119
+ + E G + T R H H IV+G+ +W G + G V K+I Y KY TK I
Sbjct 98 FLVGEYGAQ-TYRPHYHAIVFGVENIDAFLGEWRMGNVHAGT-VTAKSIYYTLKYCTKSI 155
Query 120 DEDHPEFVG------KVLCSKGIGAGYIKRADASKHKYEKGKTIETYRLRNGAKINLPIY 173
E + K L SKG+G ++ S KY K ++ L G I LP Y
Sbjct 156 TEGPDKDPDDDRKPEKALMSKGLGLSHLTE---SMIKYYKDDVSRSFSLLGGTTIALPRY 212
Query 174 YRNKLFTEEER 184
YR+K+F++ E+
Sbjct 213 YRDKVFSDIEK 223
>gi|649562725|gb|KDS68909.1| hypothetical protein M096_3339 [Parabacteroides distasonis str.
3999B T(B) 6]
Length=250
Score = 66.2 bits (160), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 64/234 (27%), Positives = 107/234 (46%), Gaps = 46/234 (20%)
Query 1 MSEELRTEPNAYFMTLTISDENYEILKNTCKSED--KNTIAT---KAIRLTLERIRKKTG 55
M E P + F+TLT DE+ + ED K T+ + I+L ++R+RKK
Sbjct 1 MQAEADEYPFSLFVTLTYDDEH---IPTAMIGEDLFKTTVGVVSKRDIQLFMKRLRKKYA 57
Query 56 KSIKHWFITELGHEKTERLHLHGIVWGI------GTDQLIKEKWNYGITYTGNFVNEKTI 109
+ +F+T + R H H I++G G D L+ E W G + + K I
Sbjct 58 QYRLRYFLTSEYGSQGGRPHYHMILFGFPFTGKHGGD-LLAECWKNGFV-QAHPLTTKEI 115
Query 110 NYITKYM------TKIDEDHPEFVGKVLCSKGIGAGYIKRADASKHKYEKGKTIETYRLR 163
+Y+TKYM I + E+ +LCSK G GY + + + ++ YRL
Sbjct 116 SYVTKYMYEKSMIPDILKGVKEYQPFMLCSKMPGIGY---------HFLREQILDFYRLH 166
Query 164 --------NGAKINLPIYYRNKLFTE-------EERELLFIDKIEKGFIYVLGT 202
NG ++ +P YY +KL+ + E RE FI+++++ + + + T
Sbjct 167 PRDYVRAFNGMRMAMPRYYADKLYDDDMKEYLKELREAFFINQMQQEWYHYINT 220
>gi|568293148|gb|ETN80369.1| hypothetical protein NECAME_18023 [Necator americanus]
Length=345
Score = 67.0 bits (162), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 57/200 (29%), Positives = 93/200 (47%), Gaps = 33/200 (17%)
Query 1 MSEELRTEPNAYFMTLTISDENYEILKNTCKSEDKNTIATKAIRLTLERIRK-KTGKSIK 59
+ EEL+ E NA F+TLT I KN + D+ ++R+RK G+ +K
Sbjct 41 LQEELQHE-NASFVTLTYDTRFVPISKNGFMTLDRGEFPR-----YMKRLRKLVPGRKLK 94
Query 60 HWFITELGHEKTERLHLHGIVWGIGTDQLIKEKWNYGITYTGNFVNE--------KTINY 111
++ E G ++ R H H I++G+ D L + W T G+ + K+I Y
Sbjct 95 YYMCGEYGSQRF-RPHYHAIIFGVPQDSLFADAW----TLNGDSLGGVVVGTVTGKSIAY 149
Query 112 ITKYMTKI--------DEDHPEFVGKVLCSKGIGAGYIKRADASKHKYEKGKTIETYRLR 163
KY+ K D+ PEF L SKG+G Y+ HK + + T
Sbjct 150 TMKYIDKSTWKQKHGRDDRVPEFS---LMSKGMGVSYLTPQMVEYHKEDISRLFCTRE-- 204
Query 164 NGAKINLPIYYRNKLFTEEE 183
G++I +P YYR K++++++
Sbjct 205 GGSRIAMPRYYRQKIYSDDD 224
>gi|492501778|ref|WP_005867316.1| hypothetical protein [Parabacteroides distasonis]
gi|409230407|gb|EKN23271.1| hypothetical protein HMPREF1059_03256 [Parabacteroides distasonis
CL09T03C24]
Length=284
Score = 65.1 bits (157), Expect = 5e-09, Method: Compositional matrix adjust.
Identities = 59/220 (27%), Positives = 102/220 (46%), Gaps = 50/220 (23%)
Query 9 PNAYFMTLTISDENY-------EILKNTCKSEDKNTIATKAIRLTLERIRKKTGKSIKHW 61
P + F+TLT DE+ ++ K+T ++ + I+L ++R+RKK + +
Sbjct 43 PFSLFVTLTYDDEHMPTAMIGEDLFKSTV-----GVVSKRDIQLFMKRLRKKYDQYRLRY 97
Query 62 FITELGHEKTERLHLHGIVWGI------GTDQLIKEKWNYGITYTGNFVNEKTINYITKY 115
F+T + R H H I++G G D L+ E W G + + K I Y+TKY
Sbjct 98 FLTSEYGSQGGRPHYHMILFGFPFTGKHGGD-LLAECWKNGFV-QAHPLTTKEIAYVTKY 155
Query 116 M------TKIDEDHPEFVGKVLCSKGIGAGYIKRADASKHKYEKGKTIETYRLR------ 163
M I +D E+ +LCS+ G GY + + + ++ YRL
Sbjct 156 MYEKSMVPDILKDVKEYQPFMLCSRIPGIGY---------HFLREQILDFYRLHPRDYVR 206
Query 164 --NGAKINLPIYYRNKLFTE-------EERELLFIDKIEK 194
NG ++ +P YY +KL+ + E RE FI+++++
Sbjct 207 AFNGMRMAMPRYYADKLYDDDMKEYLKELREAFFINQMQQ 246
>gi|649555288|gb|KDS61825.1| hypothetical protein M095_3808 [Parabacteroides distasonis str.
3999B T(B) 4]
gi|649560564|gb|KDS66872.1| hypothetical protein M095_2449 [Parabacteroides distasonis str.
3999B T(B) 4]
gi|649561011|gb|KDS67298.1| hypothetical protein M095_2409 [Parabacteroides distasonis str.
3999B T(B) 4]
Length=284
Score = 65.1 bits (157), Expect = 5e-09, Method: Compositional matrix adjust.
Identities = 60/228 (26%), Positives = 105/228 (46%), Gaps = 50/228 (22%)
Query 9 PNAYFMTLTISDENY-------EILKNTCKSEDKNTIATKAIRLTLERIRKKTGKSIKHW 61
P + F+TLT DE+ ++ K T ++ + I+L ++R+RKK + +
Sbjct 43 PFSLFVTLTYDDEHIPTAMIGEDLFKTTV-----GVVSKRDIQLFMKRLRKKYAQYRLRY 97
Query 62 FITELGHEKTERLHLHGIVWGI------GTDQLIKEKWNYGITYTGNFVNEKTINYITKY 115
F+T + R H H I++G G D L+ E W G + + K I+Y+TKY
Sbjct 98 FLTSEYGSQGGRPHYHMILFGFPFTGKHGGD-LLAECWKNGFV-QAHPLTTKEISYVTKY 155
Query 116 M------TKIDEDHPEFVGKVLCSKGIGAGYIKRADASKHKYEKGKTIETYRLR------ 163
M I + E+ +LCSK G GY + + + ++ YRL
Sbjct 156 MYEKSMIPDILKGVKEYQPFMLCSKMPGIGY---------HFLREQILDFYRLHPRDYVR 206
Query 164 --NGAKINLPIYYRNKLFTE-------EERELLFIDKIEKGFIYVLGT 202
NG ++ +P YY +KL+ + E RE FI+++++ + + + T
Sbjct 207 AFNGMRMAMPRYYADKLYDDDMKEYLKELREAFFINQMQQEWYHYINT 254
>gi|547920048|ref|WP_022322419.1| putative replication protein [Parabacteroides merdae CAG:48]
gi|524592960|emb|CDD13572.1| putative replication protein [Parabacteroides merdae CAG:48]
Length=278
Score = 62.4 bits (150), Expect = 4e-08, Method: Compositional matrix adjust.
Identities = 52/202 (26%), Positives = 94/202 (47%), Gaps = 19/202 (9%)
Query 1 MSEELRTEPNAYFMTLTISDENYEI--LKNTCKSEDKNTIATKAIRLTLERIRKKTGKSI 58
+ E + P + F+TLT DE+ I + + + ++ + ++L ++R+RKK
Sbjct 30 LQAEAKEYPLSLFVTLTYDDEHLPIERIGSDLFQTNVAVVSKRDVQLFMKRLRKKYEDYK 89
Query 59 KHWFITELGHEKTERLHLHGIVWGIG-----TDQLIKEKWNYGITYTGNFVNEKTINYIT 113
+F+T K R H H I++G L+ E W G + + K I Y+
Sbjct 90 MRYFVTSEYGAKNGRPHYHMILFGFPFTGKMAGDLLAECWQNGFV-QAHPLTIKEIAYVC 148
Query 114 KYM------TKIDEDHPEFVGKVLCSK--GIGAGYIKRADASKHKYEKGKTIETYRLRNG 165
KYM +I D ++ +LCS+ GIG G++K A ++ + + R G
Sbjct 149 KYMYEKSMCPEILRDEKKYKPFMLCSRNPGIGFGFMK---ADIIEFYRRHPRDYVRAWAG 205
Query 166 AKINLPIYYRNKLFTEEERELL 187
K+ +P YY +KL+ ++ + L
Sbjct 206 HKMAMPRYYADKLYDDDMKAFL 227
>gi|575094608|emb|CDL65959.1| unnamed protein product [uncultured bacterium]
Length=251
Score = 54.7 bits (130), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 45/149 (30%), Positives = 70/149 (47%), Gaps = 35/149 (23%)
Query 13 FMTLTISDEN-----YEILKNTCKSEDKNTIATKAIRLTLERIRKKTG-KSIKHWFITEL 66
F+TLT +D+N + L + CK + ++L ++R+RK K I+ + E
Sbjct 8 FITLTYNDDNLPYDVFSPLPSLCKRD---------VQLFMKRLRKMFSYKQIRFYLCGEY 58
Query 67 GHEKTERLHLHGIVWGI---------GTDQLIKEKWNYGITYTGNFVNEKTINYITKYMT 117
G E+T R H H I++G G+ + ++ W +G Y G N KTI Y+ Y+T
Sbjct 59 G-EQTHRPHYHAIIFGHDFNADTDFHGSSKTLEHLWQFGNNYVGQ-CNPKTIQYVAGYVT 116
Query 118 ------KIDEDHPEFVGKVLCSKGIGAGY 140
K D PEF L S+ G G+
Sbjct 117 KKYVNKKRDTITPEF---TLMSRRPGIGF 142
>gi|497292362|ref|WP_009606579.1| hypothetical protein [Turicibacter sp. HGF1]
gi|325490295|gb|EGC92624.1| hypothetical protein HMPREF9402_0015 [Turicibacter sp. HGF1]
Length=309
Score = 47.0 bits (110), Expect = 0.007, Method: Compositional matrix adjust.
Identities = 48/145 (33%), Positives = 72/145 (50%), Gaps = 9/145 (6%)
Query 47 LERIRKKTG-KSIKHWFITELGHEKTERLHLHGIV-WGIGTDQLIKEKWNYGIT--YTGN 102
+ R+RKK G ++ K+ ++ E G TER HLH I+ G+ D+ I+EKW G T T N
Sbjct 134 INRLRKKKGLENAKYVYVIEEGTFGTERFHLHLIIDNGLSKDE-IEEKWGLGATTLRTLN 192
Query 103 FVNEKTINYITKYMTKIDEDHPEFVGKVLCSK--GIGAGYIKRADASKH--KYEKGKTIE 158
+ E+ + KYM K +E + ++ + G G +K ASK+ K K K +E
Sbjct 193 YYKEENFIGVCKYMMKDEETYKRTAFRLKGKRRWGSSKGNLKVPKASKNRTKMSKKKVME 252
Query 159 TYRLRNGAKINLPIYYRNKLFTEEE 183
++G L Y N F E E
Sbjct 253 MVLHQDGIGEKLEREYFNHQFKEVE 277
>gi|489684024|ref|WP_003588246.1| MULTISPECIES: hypothetical protein [Lactobacillus casei group]
gi|410534632|gb|EKQ09274.1| hypothetical protein LCAM36_0721 [Lactobacillus casei M36]
gi|410546224|gb|EKQ20487.1| hypothetical protein LCAUW1_1935 [Lactobacillus casei UW1]
gi|511394685|gb|EPC32972.1| Rep protein [Lactobacillus paracasei subsp. paracasei Lpp22]
Length=290
Score = 44.3 bits (103), Expect = 0.050, Method: Compositional matrix adjust.
Identities = 42/135 (31%), Positives = 59/135 (44%), Gaps = 32/135 (24%)
Query 5 LRTEPNAYFMTLTISDENYEILKNTCKSEDKNTI-ATKAIRLTLERIRKKTGKSIKHWFI 63
L P F TLT D K KN + A +R L+ R+K G+ + FI
Sbjct 72 LEANPFNLFWTLTFDD---------SKVNAKNYVYARNRLRAWLKYQREKFGR-FDYLFI 121
Query 64 TELGHEKTERLHLHGIVWGIG----------TDQLIKEK---------WNYGITYTGNFV 104
EL H T R+H HG+ G+ + +LIK+K W G + +
Sbjct 122 PEL-HPSTGRIHFHGVTGGLAPPLTPARYPKSQRLIKKKGLQIYNADSWEKGFSTVSHIA 180
Query 105 NE-KTINYITKYMTK 118
++ K NYITKY+TK
Sbjct 181 DKRKAANYITKYITK 195
Lambda K H a alpha
0.317 0.137 0.407 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 1363403997231