bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: All non-redundant GenBank CDS translations+PDB+SwissProt+PIR+PRF
excluding environmental samples from WGS projects
49,011,213 sequences; 17,563,301,199 total letters
Query= Contig-7_CDS_annotation_glimmer3.pl_2_7
Length=106
Score E
Sequences producing significant alignments: (Bits) Value
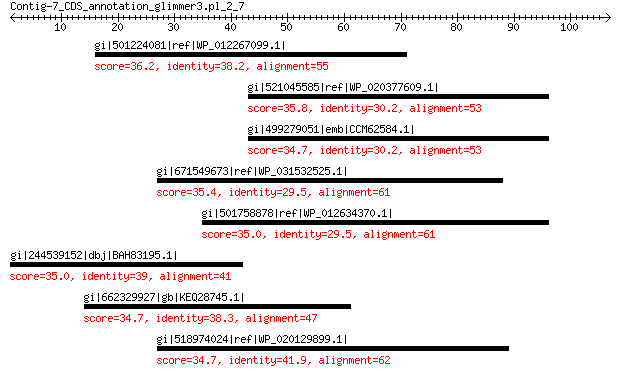
gi|501224081|ref|WP_012267099.1| hypothetical protein 36.2 0.38
gi|521045585|ref|WP_020377609.1| hypothetical protein 35.8 1.1
gi|499279051|emb|CCM62584.1| putative growth inhibitor 34.7 2.5
gi|671549673|ref|WP_031532525.1| MULTISPECIES: hypothetical protein 35.4 3.4
gi|501758878|ref|WP_012634370.1| hypothetical protein 35.0 4.8
gi|244539152|dbj|BAH83195.1| DNA-binding ATP-dependent protease La 35.0 5.4
gi|662329927|gb|KEQ28745.1| hypothetical protein N180_18775 34.7 8.0
gi|518974024|ref|WP_020129899.1| histidinol dehydrogenase 34.7 8.6
>gi|501224081|ref|WP_012267099.1| hypothetical protein [Microcystis aeruginosa]
gi|166367241|ref|YP_001659514.1| hypothetical protein MAE_45000 [Microcystis aeruginosa NIES-843]
gi|166089614|dbj|BAG04322.1| unknown protein [Microcystis aeruginosa NIES-843]
Length=55
Score = 36.2 bits (82), Expect = 0.38, Method: Compositional matrix adjust.
Identities = 21/55 (38%), Positives = 31/55 (56%), Gaps = 5/55 (9%)
Query 16 SVECFEGEQIEEKVRRIVNNNEPITDGAPIIFTEKKDGVRPEYNVRTDRWDIALD 70
S+ GE + +K +R +NN IT G IIF EK PE++ R+ +W + LD
Sbjct 2 SIGYISGEALMQKAKRRINNQFLIT-GFSIIFREKV----PEFSPRSLQWPVLLD 51
>gi|521045585|ref|WP_020377609.1| hypothetical protein [Candidatus Microthrix parvicella]
Length=109
Score = 35.8 bits (81), Expect = 1.1, Method: Compositional matrix adjust.
Identities = 16/53 (30%), Positives = 26/53 (49%), Gaps = 0/53 (0%)
Query 43 APIIFTEKKDGVRPEYNVRTDRWDIALDAMNKIDMARKAHKENEVKPEDFGNV 95
AP + + RPE VR +R + ++ M +D+AR E + PE+ V
Sbjct 42 APTSLSARPASYRPEVEVRGERTRVLVEQMAAVDVARLGELEGHLGPEELWGV 94
>gi|499279051|emb|CCM62584.1| putative growth inhibitor [Candidatus Microthrix parvicella RN1]
Length=109
Score = 34.7 bits (78), Expect = 2.5, Method: Compositional matrix adjust.
Identities = 16/53 (30%), Positives = 25/53 (47%), Gaps = 0/53 (0%)
Query 43 APIIFTEKKDGVRPEYNVRTDRWDIALDAMNKIDMARKAHKENEVKPEDFGNV 95
AP + + RPE VR R + ++ M +D+AR E + PE+ V
Sbjct 42 APTSLSARPASYRPEVEVRGARTRVLVEQMAAVDVARLGELEGHLGPEELWGV 94
>gi|671549673|ref|WP_031532525.1| MULTISPECIES: hypothetical protein [Bacteroides]
Length=313
Score = 35.4 bits (80), Expect = 3.4, Method: Compositional matrix adjust.
Identities = 18/61 (30%), Positives = 34/61 (56%), Gaps = 0/61 (0%)
Query 27 EKVRRIVNNNEPITDGAPIIFTEKKDGVRPEYNVRTDRWDIALDAMNKIDMARKAHKENE 86
E + +++ NN+ I D A I K G+R ++ + D ++ ++A +I M KA +E E
Sbjct 88 EDLNKLILNNQDINDEAAIAEVSKAMGLRDKFQTKIDEYNAWVEAKREIAMTAKAKREAE 147
Query 87 V 87
+
Sbjct 148 L 148
>gi|501758878|ref|WP_012634370.1| hypothetical protein [Methylobacterium nodulans]
gi|220920133|ref|YP_002495434.1| hypothetical protein Mnod_0081 [Methylobacterium nodulans ORS
2060]
gi|219944739|gb|ACL55131.1| hypothetical protein Mnod_0081 [Methylobacterium nodulans ORS
2060]
Length=198
Score = 35.0 bits (79), Expect = 4.8, Method: Compositional matrix adjust.
Identities = 18/65 (28%), Positives = 29/65 (45%), Gaps = 4/65 (6%)
Query 35 NNEPITDGAPIIFTEKKDGVRPEYNVRTDRWDIALDAMNKIDMARKAHK----ENEVKPE 90
N E I ++ +D R + + RW LD +NK+ AR H+ ++EV E
Sbjct 102 NAEAIARCGTVLTQAVQDAFRNSFELGQKRWQSNLDGLNKLTRARSVHELAMLQSEVARE 161
Query 91 DFGNV 95
N+
Sbjct 162 SLQNL 166
>gi|244539152|dbj|BAH83195.1| DNA-binding ATP-dependent protease La [Candidatus Ishikawaella
capsulata Mpkobe]
Length=793
Score = 35.0 bits (79), Expect = 5.4, Method: Compositional matrix adjust.
Identities = 16/41 (39%), Positives = 22/41 (54%), Gaps = 0/41 (0%)
Query 1 MYKKTIIPGYTGRMKSVECFEGEQIEEKVRRIVNNNEPITD 41
+Y +IP + GR KS+ C E EEK +V EP+ D
Sbjct 35 VYPHMVIPLFVGREKSIRCLESAVEEEKKILLVAQREPLQD 75
>gi|662329927|gb|KEQ28745.1| hypothetical protein N180_18775 [Sphingobacterium antarcticus
4BY]
Length=315
Score = 34.7 bits (78), Expect = 8.0, Method: Compositional matrix adjust.
Identities = 18/47 (38%), Positives = 28/47 (60%), Gaps = 3/47 (6%)
Query 14 MKSVECFEGEQIEEKVRRIVNNNEPITDGAPIIFTEKKDGVRPEYNV 60
+K+V+ FE + E +RR+ NN + + DG F KK+G EYN+
Sbjct 79 IKAVQKFENKSFVEAIRRLSNNTDIVADGYQAFF--KKNG-ETEYNI 122
>gi|518974024|ref|WP_020129899.1| histidinol dehydrogenase [Streptomyces sp. 303MFCol5.2]
Length=441
Score = 34.7 bits (78), Expect = 8.6, Method: Composition-based stats.
Identities = 26/62 (42%), Positives = 34/62 (55%), Gaps = 2/62 (3%)
Query 27 EKVRRIVNNNEPITDGAPIIFTEKKDGVRPEYNVRTDRWDIALDAMNKIDMARKAHKENE 86
EKVR I D A I F EK DGVR E +VR IA DA++++D A +A E
Sbjct 34 EKVRPICEAVHHRGDAALIDFAEKFDGVRLE-SVRVPAAAIA-DALDRLDPAVRAALEES 91
Query 87 VK 88
++
Sbjct 92 IR 93
Lambda K H a alpha
0.312 0.133 0.388 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 430511266944