bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: All non-redundant GenBank CDS translations+PDB+SwissProt+PIR+PRF
excluding environmental samples from WGS projects
49,011,213 sequences; 17,563,301,199 total letters
Query= Contig-8_CDS_annotation_glimmer3.pl_2_4
Length=54
Score E
Sequences producing significant alignments: (Bits) Value
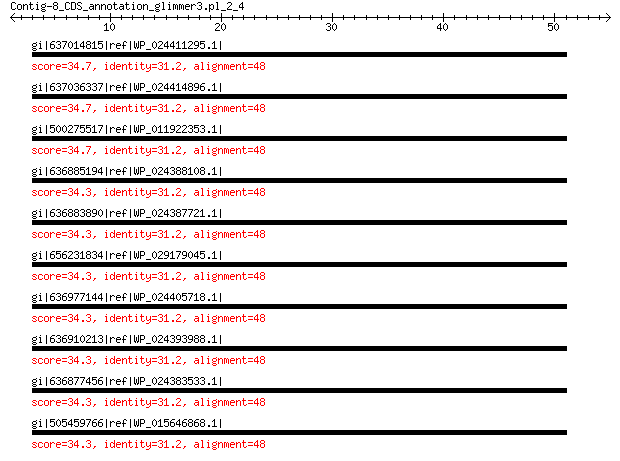
gi|637014815|ref|WP_024411295.1| hypothetical protein 34.7 2.5
gi|637036337|ref|WP_024414896.1| hypothetical protein 34.7 2.6
gi|500275517|ref|WP_011922353.1| hypothetical protein 34.7 2.6
gi|636885194|ref|WP_024388108.1| hypothetical protein 34.3 2.8
gi|636883890|ref|WP_024387721.1| hypothetical protein 34.3 2.8
gi|656231834|ref|WP_029179045.1| hypothetical protein 34.3 2.9
gi|636977144|ref|WP_024405718.1| hypothetical protein 34.3 2.9
gi|636910213|ref|WP_024393988.1| hypothetical protein 34.3 2.9
gi|636877456|ref|WP_024383533.1| hypothetical protein 34.3 2.9
gi|505459766|ref|WP_015646868.1| DegV family protein 34.3 2.9
>gi|637014815|ref|WP_024411295.1| hypothetical protein [Streptococcus suis]
Length=283
Score = 34.7 bits (78), Expect = 2.5, Method: Compositional matrix adjust.
Identities = 15/48 (31%), Positives = 30/48 (63%), Gaps = 0/48 (0%)
Query 3 KGVLKFIKRETLHEEFVISIGIFNRPSTAERFRKQLQDVNVGYDVLLI 50
K +L+ ++ +T ++ I+I N P AE F++QL++ VG D+ ++
Sbjct 212 KRLLEILQEQTTEGQYQIAIIHANSPQKAENFKQQLEEAGVGSDLPIV 259
>gi|637036337|ref|WP_024414896.1| hypothetical protein [Streptococcus suis]
Length=283
Score = 34.7 bits (78), Expect = 2.6, Method: Compositional matrix adjust.
Identities = 15/48 (31%), Positives = 30/48 (63%), Gaps = 0/48 (0%)
Query 3 KGVLKFIKRETLHEEFVISIGIFNRPSTAERFRKQLQDVNVGYDVLLI 50
K +L+ ++ +T ++ I+I N P AE F++QL++ VG D+ ++
Sbjct 212 KRLLEILQEQTTEGQYQIAIIHANSPQKAENFKQQLEEAGVGSDLPIV 259
>gi|500275517|ref|WP_011922353.1| hypothetical protein [Streptococcus suis]
gi|146318481|ref|YP_001198193.1| hypothetical protein SSU05_0827 [Streptococcus suis 05ZYH33]
gi|146320674|ref|YP_001200385.1| hypothetical protein SSU98_0827 [Streptococcus suis 98HAH33]
gi|253751608|ref|YP_003024749.1| DegV family protein [Streptococcus suis SC84]
gi|253753510|ref|YP_003026651.1| DegV family protein [Streptococcus suis P1/7]
gi|253755665|ref|YP_003028805.1| DegV family protein [Streptococcus suis BM407]
gi|386577800|ref|YP_006074206.1| DegV [Streptococcus suis GZ1]
gi|386579855|ref|YP_006076260.1| hypothetical protein SSUJS14_0908 [Streptococcus suis JS14]
gi|386581800|ref|YP_006078204.1| hypothetical protein SSU12_0769 [Streptococcus suis SS12]
gi|386588034|ref|YP_006084435.1| hypothetical protein SSUA7_0766 [Streptococcus suis A7]
gi|403061438|ref|YP_006649654.1| hypothetical protein YYK_03685 [Streptococcus suis S735]
gi|476419821|ref|YP_007570817.1| DegV domain-containing protein SP [Streptococcus suis SC070731]
gi|145689287|gb|ABP89793.1| Uncharacterized protein conserved in bacteria [Streptococcus
suis 05ZYH33]
gi|145691480|gb|ABP91985.1| Uncharacterized protein conserved in bacteria [Streptococcus
suis 98HAH33]
gi|251815897|emb|CAZ51511.1| DegV family protein [Streptococcus suis SC84]
gi|251818129|emb|CAZ55923.1| DegV family protein [Streptococcus suis BM407]
gi|251819756|emb|CAR45644.1| DegV family protein [Streptococcus suis P1/7]
gi|292558263|gb|ADE31264.1| DegV [Streptococcus suis GZ1]
gi|319758047|gb|ADV69989.1| hypothetical protein SSUJS14_0908 [Streptococcus suis JS14]
gi|353733946|gb|AER14956.1| hypothetical protein SSU12_0769 [Streptococcus suis SS12]
gi|354985195|gb|AER44093.1| hypothetical protein SSUA7_0766 [Streptococcus suis A7]
gi|402808764|gb|AFR00256.1| hypothetical protein YYK_03685 [Streptococcus suis S735]
gi|459386628|gb|AGG64336.1| DegV domain-containing protein SP [Streptococcus suis SC070731]
Length=283
Score = 34.7 bits (78), Expect = 2.6, Method: Compositional matrix adjust.
Identities = 15/48 (31%), Positives = 30/48 (63%), Gaps = 0/48 (0%)
Query 3 KGVLKFIKRETLHEEFVISIGIFNRPSTAERFRKQLQDVNVGYDVLLI 50
K +L+ ++ +T ++ I+I N P AE F++QL++ VG D+ ++
Sbjct 212 KRLLEILQEQTTEGQYQIAIIHANAPQKAENFKRQLEEAGVGSDLPIV 259
>gi|636885194|ref|WP_024388108.1| hypothetical protein [Streptococcus suis]
Length=283
Score = 34.3 bits (77), Expect = 2.8, Method: Compositional matrix adjust.
Identities = 15/48 (31%), Positives = 30/48 (63%), Gaps = 0/48 (0%)
Query 3 KGVLKFIKRETLHEEFVISIGIFNRPSTAERFRKQLQDVNVGYDVLLI 50
K +L+ ++ +T ++ I+I N P AE F++QL++ VG D+ ++
Sbjct 212 KRLLEILQEQTTEGQYQIAIIHANAPQKAENFKQQLEEAGVGSDLPIV 259
>gi|636883890|ref|WP_024387721.1| hypothetical protein [Streptococcus suis]
Length=283
Score = 34.3 bits (77), Expect = 2.8, Method: Compositional matrix adjust.
Identities = 15/48 (31%), Positives = 30/48 (63%), Gaps = 0/48 (0%)
Query 3 KGVLKFIKRETLHEEFVISIGIFNRPSTAERFRKQLQDVNVGYDVLLI 50
K +L+ ++ +T ++ I+I N P AE F++QL++ VG D+ ++
Sbjct 212 KRLLEILQEQTTEGQYQIAIIHANAPQKAENFKQQLEEAGVGSDLPIV 259
>gi|656231834|ref|WP_029179045.1| hypothetical protein [Streptococcus suis]
Length=283
Score = 34.3 bits (77), Expect = 2.9, Method: Compositional matrix adjust.
Identities = 15/48 (31%), Positives = 30/48 (63%), Gaps = 0/48 (0%)
Query 3 KGVLKFIKRETLHEEFVISIGIFNRPSTAERFRKQLQDVNVGYDVLLI 50
K +L+ ++ +T ++ I+I N P AE F++QL++ VG D+ ++
Sbjct 212 KRLLEILQEQTTEGQYQIAIIHANAPQKAENFKQQLEEAGVGSDLPIV 259
>gi|636977144|ref|WP_024405718.1| hypothetical protein [Streptococcus suis]
Length=283
Score = 34.3 bits (77), Expect = 2.9, Method: Compositional matrix adjust.
Identities = 15/48 (31%), Positives = 30/48 (63%), Gaps = 0/48 (0%)
Query 3 KGVLKFIKRETLHEEFVISIGIFNRPSTAERFRKQLQDVNVGYDVLLI 50
K +L+ ++ +T ++ I+I N P AE F++QL++ VG D+ ++
Sbjct 212 KRLLEILQEQTTEGQYQIAIIHANAPQKAENFKQQLEEAGVGSDLPIV 259
>gi|636910213|ref|WP_024393988.1| hypothetical protein [Streptococcus suis]
Length=283
Score = 34.3 bits (77), Expect = 2.9, Method: Compositional matrix adjust.
Identities = 15/48 (31%), Positives = 30/48 (63%), Gaps = 0/48 (0%)
Query 3 KGVLKFIKRETLHEEFVISIGIFNRPSTAERFRKQLQDVNVGYDVLLI 50
K +L+ ++ +T ++ I+I N P AE F++QL++ VG D+ ++
Sbjct 212 KRLLEILQEQTTEGQYQIAIIHANAPQKAENFKQQLEEAGVGSDLPIV 259
>gi|636877456|ref|WP_024383533.1| hypothetical protein [Streptococcus suis]
Length=283
Score = 34.3 bits (77), Expect = 2.9, Method: Compositional matrix adjust.
Identities = 15/48 (31%), Positives = 30/48 (63%), Gaps = 0/48 (0%)
Query 3 KGVLKFIKRETLHEEFVISIGIFNRPSTAERFRKQLQDVNVGYDVLLI 50
K +L+ ++ +T ++ I+I N P AE F++QL++ VG D+ ++
Sbjct 212 KRLLEILQEQTTEGQYQIAIIHANAPQKAENFKQQLEEAGVGSDLPIV 259
>gi|505459766|ref|WP_015646868.1| DegV family protein [Streptococcus suis]
gi|499143281|ref|YP_007975345.1| DegV family protein [Streptococcus suis TL13]
gi|556588711|ref|YP_008756265.1| degV family protein [Streptococcus suis T15]
gi|498538158|gb|AGL47920.1| DegV family protein [Streptococcus suis TL13]
gi|556049580|gb|AGZ23225.1| degV family protein [Streptococcus suis T15]
Length=283
Score = 34.3 bits (77), Expect = 2.9, Method: Compositional matrix adjust.
Identities = 15/48 (31%), Positives = 30/48 (63%), Gaps = 0/48 (0%)
Query 3 KGVLKFIKRETLHEEFVISIGIFNRPSTAERFRKQLQDVNVGYDVLLI 50
K +L+ ++ +T ++ I+I N P AE F++QL++ VG D+ ++
Sbjct 212 KRLLEILQEQTTEGQYQIAIIHANAPQKAENFKQQLEEAGVGSDLPIV 259
Lambda K H a alpha
0.326 0.145 0.401 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 438479958096