bitscore colors: <40, 40-50 , 50-80, 80-200, >200
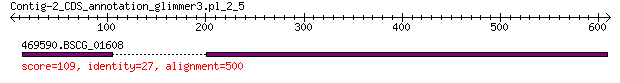
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: eggnogv4.proteins.all.fa
14,875,530 sequences; 5,112,597,290 total letters
Query= Contig-2_CDS_annotation_glimmer3.pl_2_5
Length=611
Score E
Sequences producing significant alignments: (Bits) Value
469590.BSCG_01608 109 1e-22
> 469590.BSCG_01608
Length=497
Score = 109 bits (273), Expect = 1e-22, Method: Compositional matrix adjust.
Identities = 105/415 (25%), Positives = 183/415 (44%), Gaps = 42/415 (10%)
Query 201 IPVLQSRDIELFFKRLRRNLDSHGFTSSKICYYVVSEYGPQTYRPHWHCLLFFNSEEITQ 260
+P L+ D++LF KRLR + S K+ Y+ V EYGP +RPH+H LLF S+E Q
Sbjct 117 VPYLRKTDLQLFLKRLRYYVTKQK-PSEKVRYFAVGEYGPVHFRPHYHLLLFLQSDEALQ 175
Query 261 TLREDISKAWAYGRIDYSLSRGAAASYVASYVNSAACLPFFYVGQKEIRPRSFHSKGFGS 320
E+ISKAW +GR+D +S+G ++YVASYVNS+ +P + + P S HS+ G
Sbjct 176 ICSENISKAWTFGRVDCQVSKGQCSNYVASYVNSSCTIPKVFKAS-SVCPFSVHSQKLGQ 234
Query 321 NKIFPKSSDVSEISKISDLFFDGVNVDSNGKVINIRPVRSCELAVFPRFSNDFFSDSDTC 380
+ +I ++ F ++ NGK RSC +PR
Sbjct 235 GFL---DCQREKIYSLTPENFIRSSIVLNGKYKEFDVWRSCYSFFYPR------------ 279
Query 381 CKLFQSVIETPERLVSRGYLGIDTPSF----GSDGFRLSDLVRAYSEYYERNFTDFSFSL 436
CK F + R + Y DT F L+ + Y Y+ + L
Sbjct 280 CKGF---VTKSSRERAYSYSIYDTARLLFPDAKTTFSLAKEIAIYIYYFHNPKETYLLDL 336
Query 437 RFLRGYRTRDYSDELIFREARLFDGYVFNKDMIFGRLYRLFAKVLRCFRFWNLKQYTDSW 496
+++ Y F ++ + + FN ++R++ ++L F ++
Sbjct 337 YGYCSDQSKLYELSQYFYDSDVL-LHSFNSGEFSRYVHRIYTELLISKHFLYFVCTHNTL 395
Query 497 SLKAAVKKIWSYGWEYWKKKEYRFLTTYFEYLEGCNDDERLFLLVRTSG-SGLATDSPHS 555
+ + + +++ E++ + +Y LT +FE ++LF G L TD +
Sbjct 396 AERKSKQRLIE---EFYSRLDYMHLTKFFE-------AQQLFYESDLIGDDDLCTD---N 442
Query 556 WTYTQREDYVNCLPDD--LYKRYMKTLKWLTARTEKVLKDKVKHKEFNDMQGVLL 608
W + + N + D L+++ + ++ +K+ D++KHK+ ND V
Sbjct 443 WDNSYYPYFYNNVYTDTNLFEK-TPVYRLYSSDVKKLFNDRIKHKKLNDANKVFF 496
Score = 57.4 bits (137), Expect = 6e-06, Method: Compositional matrix adjust.
Identities = 30/94 (32%), Positives = 55/94 (59%), Gaps = 3/94 (3%)
Query 13 FTECLRPQRIVNPYSHDVIFAPCGRCKSCIMNKSNFATAYAMNMATH-FKYCYFVTLTYK 71
F +CL P+RI+NPY+ + + PCG C++C + K N A+ ++ ++ K+ F+TLTY
Sbjct 6 FCKCLHPKRIMNPYTKESMVVPCGHCQACTLAK-NSRYAFQCDLESYTAKHTLFITLTYA 64
Query 72 DIFLP-YLSVEVVRRSGNRYLFDENFETMVSTSD 104
+ F+P + V+ + R L D+ ++ +D
Sbjct 65 NRFIPRAMFVDSIERPYGCDLIDKETGEILGPAD 98
Lambda K H a alpha
0.324 0.139 0.434 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 1367458008150