bitscore colors: <40, 40-50 , 50-80, 80-200, >200
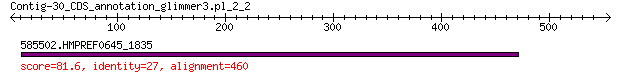
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: eggnogv4.proteins.all.fa
14,875,530 sequences; 5,112,597,290 total letters
Query= Contig-30_CDS_annotation_glimmer3.pl_2_2
Length=555
Score E
Sequences producing significant alignments: (Bits) Value
585502.HMPREF0645_1835 81.6 1e-13
> 585502.HMPREF0645_1835
Length=553
Score = 81.6 bits (200), Expect = 1e-13, Method: Compositional matrix adjust.
Identities = 124/527 (24%), Positives = 204/527 (39%), Gaps = 132/527 (25%)
Query 11 LNPTRIDMSRSTFDRNSSVKTSFNVGDIVPFYVDEVLPGDTFDIDTSKVIRMPSLLTPIM 70
+ TR + +R+ FD + + + G ++P +++P D +I+ +R + T
Sbjct 8 IKATRPNRNRNAFDLSQRHLFTAHAGMLLPVLNLDLIPHDHVEINAQDFMRTLPMNTAAF 67
Query 71 DNLYLDTYYFFVPNRIVWQHWKELM-GEN------NESAWIPTTEYEVPQITAPSGGWSI 123
++ +FFVP +W + + + G N N+S T+ +VP S S+
Sbjct 68 ASMRGVYEFFFVPYHQLWAQFDQFITGMNDFHSSANKSIQGGTSPLQVPYFNVDSVFNSL 127
Query 124 GT----------------------IADYMGV-----------PTGVSGLS----VNALPF 146
T + D +G P VSGL N F
Sbjct 128 NTGKESGSGSTDDLQYKFKYGAFRLLDLLGYGRKFDSFGTAYPDNVSGLKNNLDYNCSVF 187
Query 147 R--AYALICNEWFRDENL----CDPLNIPLTDATVAGVNTGTFVTDVAKGGLPYKAAKYR 200
R AY I +++R+ N D N D G+ V D+ K L Y+ A+
Sbjct 188 RILAYNKIYQDYYRNSNYENFDTDSFNF---DKFKGGLVDAKVVADLFK--LRYRNAQ-T 241
Query 201 DYFTSCLPAPQKSEDVTIPVSSGANYPVLSLSDIVPTPGTVPVKWNDANNVVSDAQWLLG 260
DYFT+ L S + + + D +N+ +
Sbjct 242 DYFTN-----------------------LRQSQLF----SFTTAFEDVDNIN------IA 268
Query 261 GKNYNGTITSNDISLTKTNTGPTYSAVTPINLWAVNDGSVSSATINQLRLAFQVQKLYER 320
++Y + SN T +N D S +++ LR AF V KL
Sbjct 269 PRDYVKSDGSN---------------FTRVNFGVDTDSSEGDFSVSSLRAAFAVDKLLSV 313
Query 321 DARGGTRYIEVLKSHFGVTSPDARLQRPEYLGGNRIPIVISEINQTSGTSANS------- 373
R G + + +++H+GV PD+R R YLGG + +S++ QTSGT+A
Sbjct 314 TMRAGKTFQDQMRAHYGVEIPDSRDGRVNYLGGFDSDMQVSDVTQTSGTTATEYKPEAGY 373
Query 374 ----TPQGNPSGQSRTTDVHSDFKKSFVEHGFIIGVMVA----RYDHTYQQGLERFWSRK 425
+G SG+ R + D K EHG ++ + +YD T L+ +
Sbjct 374 LGRVAGKGTGSGRGR---IVFDAK----EHGVLMCIYSLVPQIQYDCTR---LDPMVDKL 423
Query 426 GRLDYYWPVFANIGEQAVLNKEIYAQGNGTD--DEVFGYQEAWADYR 470
R DY+ P F N+G Q LN + TD + V GYQ +++Y+
Sbjct 424 DRFDYFTPEFENLGMQP-LNSSYISSFCTTDPKNPVLGYQPRYSEYK 469
Lambda K H a alpha
0.316 0.133 0.410 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 1211814630400