bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: all_orgs
14,240,465 sequences; 5,121,972,263 total letters
Query= Contig-10_CDS_annotation_glimmer3.pl_2_6
Length=111
Score E
Sequences producing significant alignments: (Bits) Value
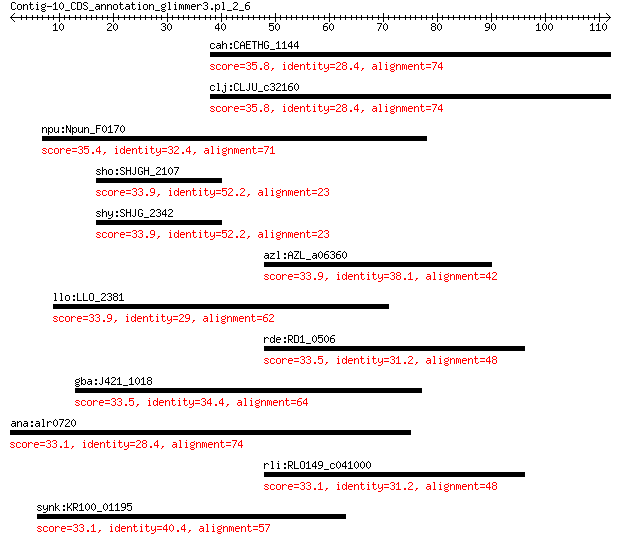
cah:CAETHG_1144 Putative amidase domain containing protein 35.8 0.62
clj:CLJU_c32160 hypothetical protein 35.8 0.62
npu:Npun_F0170 hypothetical protein 35.4 1.2
sho:SHJGH_2107 putative magnesium or manganese-dependent protein 33.9 4.9
shy:SHJG_2342 magnesium or manganese-dependent protein 33.9 4.9
azl:AZL_a06360 D-threo-aldose 1-dehydrogenase (EC:1.1.1.122) 33.9 5.2
llo:LLO_2381 coiled-coil protein 33.9 5.4
rde:RD1_0506 oxidoreductase 33.5 5.5
gba:J421_1018 Aldose 1-epimerase 33.5 5.6
ana:alr0720 hypothetical protein 33.1 7.7
rli:RLO149_c041000 pld; pyridoxal 4-dehydrogenase Pld (EC:1.1.... 33.1 8.4
synk:KR100_01195 hypothetical protein 33.1 8.7
> cah:CAETHG_1144 Putative amidase domain containing protein
Length=168
Score = 35.8 bits (81), Expect = 0.62, Method: Compositional matrix adjust.
Identities = 21/75 (28%), Positives = 40/75 (53%), Gaps = 4/75 (5%)
Query 38 TDGAPIIYTNRDDGVLPAYNIRTDRWEVAQAAMDAVNQ-ANLAKSKNYGKIEQQEQNALE 96
+ GAP++++ ++ YNIRT++W V+ ++ + KN I+ +E ++
Sbjct 52 SGGAPMVFSGKNTW---WYNIRTNKWSVSWTVAGSLYWYLKINAEKNSYGIKGREVPSIS 108
Query 97 SKEIGDTPSQQDSVG 111
S EIGD ++S G
Sbjct 109 SLEIGDLIFYRNSKG 123
> clj:CLJU_c32160 hypothetical protein
Length=168
Score = 35.8 bits (81), Expect = 0.62, Method: Compositional matrix adjust.
Identities = 21/75 (28%), Positives = 40/75 (53%), Gaps = 4/75 (5%)
Query 38 TDGAPIIYTNRDDGVLPAYNIRTDRWEVAQAAMDAVNQ-ANLAKSKNYGKIEQQEQNALE 96
+ GAP++++ ++ YNIRT++W V+ ++ + KN I+ +E ++
Sbjct 52 SGGAPMVFSGKNTW---WYNIRTNKWSVSWTVAGSLYWYLKINAEKNSYGIKGREVPSIS 108
Query 97 SKEIGDTPSQQDSVG 111
S EIGD ++S G
Sbjct 109 SLEIGDLIFYRNSKG 123
> npu:Npun_F0170 hypothetical protein
Length=231
Score = 35.4 bits (80), Expect = 1.2, Method: Compositional matrix adjust.
Identities = 23/79 (29%), Positives = 40/79 (51%), Gaps = 11/79 (14%)
Query 7 FPKPPETNYEFQDGESIENKVRRITENNEPIT-----DGAPIIYT---NRDDGVLPAYNI 58
F P E+N+ +E+ V R +++E I DG+P+I + N +GV+ + +
Sbjct 5 FFCPEESNFY---SNCLESFVIRNCKSSESIVEFGSGDGSPVINSLLRNNFNGVIQGFEL 61
Query 59 RTDRWEVAQAAMDAVNQAN 77
T W+VA + +D N N
Sbjct 62 NTSAWKVANSTIDEYNLTN 80
> sho:SHJGH_2107 putative magnesium or manganese-dependent protein
Length=690
Score = 33.9 bits (76), Expect = 4.9, Method: Composition-based stats.
Identities = 12/23 (52%), Positives = 18/23 (78%), Gaps = 0/23 (0%)
Query 17 FQDGESIENKVRRITENNEPITD 39
F D E+IE+++RR+ E EP+TD
Sbjct 193 FLDAEAIESRLRRVLETGEPVTD 215
> shy:SHJG_2342 magnesium or manganese-dependent protein
Length=690
Score = 33.9 bits (76), Expect = 4.9, Method: Composition-based stats.
Identities = 12/23 (52%), Positives = 18/23 (78%), Gaps = 0/23 (0%)
Query 17 FQDGESIENKVRRITENNEPITD 39
F D E+IE+++RR+ E EP+TD
Sbjct 193 FLDAEAIESRLRRVLETGEPVTD 215
> azl:AZL_a06360 D-threo-aldose 1-dehydrogenase (EC:1.1.1.122)
Length=353
Score = 33.9 bits (76), Expect = 5.2, Method: Composition-based stats.
Identities = 16/42 (38%), Positives = 23/42 (55%), Gaps = 0/42 (0%)
Query 48 RDDGVLPAYNIRTDRWEVAQAAMDAVNQANLAKSKNYGKIEQ 89
R DG +PA + + WEVA AMD V+ + + Y +EQ
Sbjct 189 RRDGAVPAIGLGVNEWEVALEAMDEVDLDCVMLAGRYTLLEQ 230
> llo:LLO_2381 coiled-coil protein
Length=472
Score = 33.9 bits (76), Expect = 5.4, Method: Compositional matrix adjust.
Identities = 18/65 (28%), Positives = 37/65 (57%), Gaps = 4/65 (6%)
Query 9 KPPETNYEFQDGESIENKVRRITENNEPIT---DGAPIIYTNRDDGVLPAYNIRTDRWEV 65
+PP T ++ + ES++ ++++ T + E + D I++TN+ +L RT RWE+
Sbjct 234 QPPLT-FDPKKIESLKEELQKTTNHLESVIKELDNKSIVFTNKSSKLLELQEKRTSRWEL 292
Query 66 AQAAM 70
A+ +
Sbjct 293 ARKGI 297
> rde:RD1_0506 oxidoreductase
Length=338
Score = 33.5 bits (75), Expect = 5.5, Method: Composition-based stats.
Identities = 15/48 (31%), Positives = 27/48 (56%), Gaps = 0/48 (0%)
Query 48 RDDGVLPAYNIRTDRWEVAQAAMDAVNQANLAKSKNYGKIEQQEQNAL 95
RD+GV+ A + + W+V QAA++A + + Y +EQ+ Q+
Sbjct 172 RDEGVVSAIGVGVNEWQVCQAALEARDFDCFLLAGRYTLLEQEAQDTF 219
> gba:J421_1018 Aldose 1-epimerase
Length=271
Score = 33.5 bits (75), Expect = 5.6, Method: Compositional matrix adjust.
Identities = 22/66 (33%), Positives = 29/66 (44%), Gaps = 2/66 (3%)
Query 13 TNYEFQDGESIENKVRRITENNEPITDGAPIIYTNRDD-GVLPAYNI-RTDRWEVAQAAM 70
T + DGE + R E I G P+++ D G LP + RT W+V A
Sbjct 27 TAWSTADGEQLYLSPRTAYEEGSAIRGGVPVVFPQFSDRGPLPKHGFARTRAWDVLAHAA 86
Query 71 DAVNQA 76
DAV A
Sbjct 87 DAVTLA 92
> ana:alr0720 hypothetical protein
Length=236
Score = 33.1 bits (74), Expect = 7.7, Method: Compositional matrix adjust.
Identities = 21/82 (26%), Positives = 42/82 (51%), Gaps = 12/82 (15%)
Query 1 MIKPKFFPKPPETNYEFQDGESIENKVRRITENNEPIT-----DGAPIIYT---NRDDGV 52
++K FF P E+N+ +EN + R ++ ++ I DG+P+I + N+ +G+
Sbjct 6 LMKDVFFC-PEESNFY---SNCLENFILRNSDQSDSIIEFGSGDGSPVINSLLRNQFNGI 61
Query 53 LPAYNIRTDRWEVAQAAMDAVN 74
+ + + T W+ A +D N
Sbjct 62 IHGFELNTSAWKAANLTIDEYN 83
> rli:RLO149_c041000 pld; pyridoxal 4-dehydrogenase Pld (EC:1.1.1.107)
Length=338
Score = 33.1 bits (74), Expect = 8.4, Method: Composition-based stats.
Identities = 15/48 (31%), Positives = 27/48 (56%), Gaps = 0/48 (0%)
Query 48 RDDGVLPAYNIRTDRWEVAQAAMDAVNQANLAKSKNYGKIEQQEQNAL 95
RD+GV+ A + + W+V QAA++A + + Y +EQ+ Q+
Sbjct 172 RDEGVVSAIGVGVNEWQVCQAALEARDFDCFLLAGRYTLLEQEAQDTF 219
> synk:KR100_01195 hypothetical protein
Length=761
Score = 33.1 bits (74), Expect = 8.7, Method: Composition-based stats.
Identities = 23/58 (40%), Positives = 31/58 (53%), Gaps = 3/58 (5%)
Query 6 FFPKPPETNYEFQDGES-IENKVRRITENNEPITDGAPIIYTNRDDGVLPAYNIRTDR 62
F PKP N F+DG S + K R+I EN+ I +G YT DD + P + I D+
Sbjct 627 FNPKPLLDNQLFRDGRSLLAGKSRQIPENHWSIWNG--FGYTTPDDDLRPFFRIVLDQ 682
Lambda K H a alpha
0.308 0.128 0.363 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 127903216896