bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: all_orgs
14,240,465 sequences; 5,121,972,263 total letters
Query= Contig-11_CDS_annotation_glimmer3.pl_2_3
Length=497
Score E
Sequences producing significant alignments: (Bits) Value
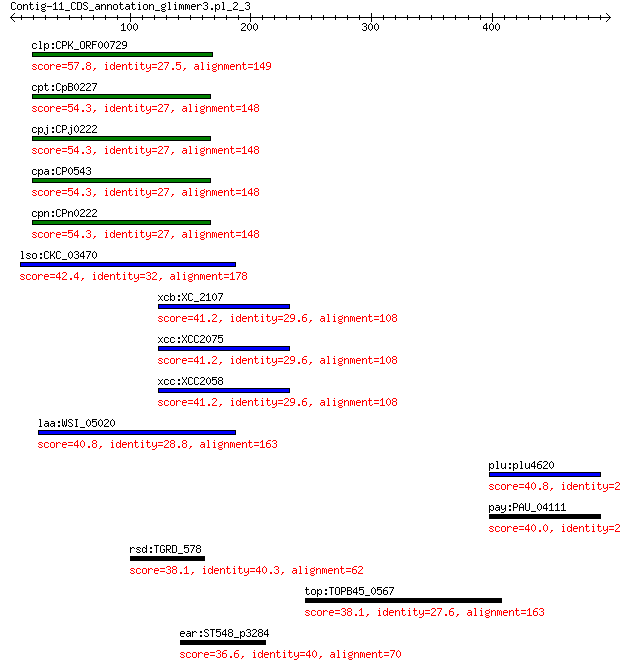
clp:CPK_ORF00729 hypothetical protein 57.8 3e-07
cpt:CpB0227 hypothetical protein 54.3 3e-06
cpj:CPj0222 hypothetical protein 54.3 3e-06
cpa:CP0543 hypothetical protein 54.3 3e-06
cpn:CPn0222 hypothetical protein 54.3 3e-06
lso:CKC_03470 hypothetical protein 42.4 0.18
xcb:XC_2107 replication initiation protein 41.2 0.37
xcc:XCC2075 gII; replication initiation protein 41.2 0.37
xcc:XCC2058 gII; replication initiation protein 41.2 0.44
laa:WSI_05020 hypothetical protein 40.8 0.54
plu:plu4620 recQ; ATP-dependent DNA helicase RecQ 40.8 0.80
pay:PAU_04111 recQ; ATP-dependent DNA helicase (EC:3.6.1.-) 40.0 1.2
rsd:TGRD_578 hypothetical protein 38.1 2.0
top:TOPB45_0567 hypothetical protein 38.1 5.4
ear:ST548_p3284 FIG00947185: hypothetical protein 36.6 8.2
> clp:CPK_ORF00729 hypothetical protein
Length=121
Score = 57.8 bits (138), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 41/149 (28%), Positives = 64/149 (43%), Gaps = 50/149 (34%)
Query 19 YTNESMVVPCGHCQACMLAKNSRYAFQCDLESYVAKHTLFITLTYANRYIPRATFVDSLE 78
Y N +++PC C+ C ++++C E+ + + F+TLTY ++++P+
Sbjct 15 YRNRWVLMPCLKCRFCRTQHAKVWSYRCVHEASLYEKNCFLTLTYDDKHLPQ-------- 66
Query 79 RPFGNDLVDKETGEILGPSDMKQEDIDRLLNKFYLFGDVPYLRKTDLQLFFKRLRYYVSK 138
+G+ L K LQLF KRLR +S
Sbjct 67 --YGS------------------------------------LVKLHLQLFLKRLRDRIS- 87
Query 139 QCPSEKVRYFAVGEYGPVHFRPHYHILLF 167
K+RYF GEYG RPHYH+L+F
Sbjct 88 ---PHKIRYFGCGEYGTKLQRPHYHLLIF 113
> cpt:CpB0227 hypothetical protein
Length=113
Score = 54.3 bits (129), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 40/148 (27%), Positives = 62/148 (42%), Gaps = 50/148 (34%)
Query 19 YTNESMVVPCGHCQACMLAKNSRYAFQCDLESYVAKHTLFITLTYANRYIPRATFVDSLE 78
Y N +++PC C+ C ++++C E+ + + F+TLTY ++++P+
Sbjct 15 YRNRWVLMPCLKCRFCRTQHAKVWSYRCVHEASLYEKNCFLTLTYDDKHLPQ-------- 66
Query 79 RPFGNDLVDKETGEILGPSDMKQEDIDRLLNKFYLFGDVPYLRKTDLQLFFKRLRYYVSK 138
+G+ L K LQLF KRLR +S
Sbjct 67 --YGS------------------------------------LVKLHLQLFLKRLRKMIS- 87
Query 139 QCPSEKVRYFAVGEYGPVHFRPHYHILL 166
K+RYF G YG RPHYH+LL
Sbjct 88 ---PHKIRYFECGAYGTKLQRPHYHLLL 112
> cpj:CPj0222 hypothetical protein
Length=113
Score = 54.3 bits (129), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 40/148 (27%), Positives = 62/148 (42%), Gaps = 50/148 (34%)
Query 19 YTNESMVVPCGHCQACMLAKNSRYAFQCDLESYVAKHTLFITLTYANRYIPRATFVDSLE 78
Y N +++PC C+ C ++++C E+ + + F+TLTY ++++P+
Sbjct 15 YRNRWVLMPCLKCRFCRTQHAKVWSYRCVHEASLYEKNCFLTLTYDDKHLPQ-------- 66
Query 79 RPFGNDLVDKETGEILGPSDMKQEDIDRLLNKFYLFGDVPYLRKTDLQLFFKRLRYYVSK 138
+G+ L K LQLF KRLR +S
Sbjct 67 --YGS------------------------------------LVKLHLQLFLKRLRKMIS- 87
Query 139 QCPSEKVRYFAVGEYGPVHFRPHYHILL 166
K+RYF G YG RPHYH+LL
Sbjct 88 ---PHKIRYFECGAYGTKLQRPHYHLLL 112
> cpa:CP0543 hypothetical protein
Length=113
Score = 54.3 bits (129), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 40/148 (27%), Positives = 62/148 (42%), Gaps = 50/148 (34%)
Query 19 YTNESMVVPCGHCQACMLAKNSRYAFQCDLESYVAKHTLFITLTYANRYIPRATFVDSLE 78
Y N +++PC C+ C ++++C E+ + + F+TLTY ++++P+
Sbjct 15 YRNRWVLMPCLKCRFCRTQHAKVWSYRCVHEASLYEKNCFLTLTYDDKHLPQ-------- 66
Query 79 RPFGNDLVDKETGEILGPSDMKQEDIDRLLNKFYLFGDVPYLRKTDLQLFFKRLRYYVSK 138
+G+ L K LQLF KRLR +S
Sbjct 67 --YGS------------------------------------LVKLHLQLFLKRLRKMIS- 87
Query 139 QCPSEKVRYFAVGEYGPVHFRPHYHILL 166
K+RYF G YG RPHYH+LL
Sbjct 88 ---PHKIRYFECGAYGTKLQRPHYHLLL 112
> cpn:CPn0222 hypothetical protein
Length=113
Score = 54.3 bits (129), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 40/148 (27%), Positives = 62/148 (42%), Gaps = 50/148 (34%)
Query 19 YTNESMVVPCGHCQACMLAKNSRYAFQCDLESYVAKHTLFITLTYANRYIPRATFVDSLE 78
Y N +++PC C+ C ++++C E+ + + F+TLTY ++++P+
Sbjct 15 YRNRWVLMPCLKCRFCRTQHAKVWSYRCVHEASLYEKNCFLTLTYDDKHLPQ-------- 66
Query 79 RPFGNDLVDKETGEILGPSDMKQEDIDRLLNKFYLFGDVPYLRKTDLQLFFKRLRYYVSK 138
+G+ L K LQLF KRLR +S
Sbjct 67 --YGS------------------------------------LVKLHLQLFLKRLRKMIS- 87
Query 139 QCPSEKVRYFAVGEYGPVHFRPHYHILL 166
K+RYF G YG RPHYH+LL
Sbjct 88 ---PHKIRYFECGAYGTKLQRPHYHLLL 112
> lso:CKC_03470 hypothetical protein
Length=424
Score = 42.4 bits (98), Expect = 0.18, Method: Compositional matrix adjust.
Identities = 57/203 (28%), Positives = 86/203 (42%), Gaps = 41/203 (20%)
Query 9 CLHPKKIVNPYTNESMVVPCGH-----------CQACMLAKNSRYAF---QCDLESYVAK 54
C +P +V YT+ ++ G+ C+ C + SR F + E +
Sbjct 194 CCNPVVLVMVYTDIVVIFDIGYKYNYFFYILARCRRCSVCCKSRGMFWLRRAQTEVMRSS 253
Query 55 HTLFITLTYA------NRYIPRATFVDSLERPFGNDLVDKET-GEILGPSDMKQEDIDRL 107
T FITLT++ N + +V+SL N K+ G I+ D++ +I +
Sbjct 254 RTWFITLTFSPSNHIKNYALTIGQYVESLSIEDRNFFYGKKKYGTII--EDIRSLNISDV 311
Query 108 LNKFYL----FGDVPYLRKTDLQLFFKRLRYYVSKQCPSEKVRYFAVGEYGPVHFRPHYH 163
KF L FGD + LF KRLR + S+K RYF V E PH H
Sbjct 312 DLKFRLLCKGFGD-------KIVLFLKRLR-----KNTSKKFRYFIVFEKHKSG-NPHAH 358
Query 164 ILLFLQSDEALQVCSENISQAWT 186
+L+ +S E L +E I + W
Sbjct 359 MLIHQKSGEELLKKAE-IQEEWI 380
> xcb:XC_2107 replication initiation protein
Length=346
Score = 41.2 bits (95), Expect = 0.37, Method: Compositional matrix adjust.
Identities = 32/120 (27%), Positives = 53/120 (44%), Gaps = 17/120 (14%)
Query 124 DLQLFFKRLRYYVSK------QCPSEKVRYFAVGEYGPVHFRPHYHILLFLQSDEALQVC 177
D+ FKR+R + ++ + E RY VGE FRPHYH++L++ + +
Sbjct 174 DVSELFKRMRGHFNRLKSGRARWNRESFRYVWVGEL-TQRFRPHYHVMLWV--PQGMFFG 230
Query 178 SENISQAWTLGRIDCQISKGQ-CSSYVASYVN-----SSCTIPKVFKLSSVCPFNVHSQK 231
+ W G QI K + C Y+A Y + ++ PK F+ N S++
Sbjct 231 KVDQRGWWPHG--SSQIEKARNCVGYLAKYASKFTAITAAAFPKGFRTHGCGGLNTESKR 288
> xcc:XCC2075 gII; replication initiation protein
Length=346
Score = 41.2 bits (95), Expect = 0.37, Method: Compositional matrix adjust.
Identities = 32/120 (27%), Positives = 53/120 (44%), Gaps = 17/120 (14%)
Query 124 DLQLFFKRLRYYVSK------QCPSEKVRYFAVGEYGPVHFRPHYHILLFLQSDEALQVC 177
D+ FKR+R + ++ + E RY VGE FRPHYH++L++ + +
Sbjct 174 DVSELFKRMRGHFNRLKSGRARWNRESFRYVWVGEL-TQRFRPHYHVMLWV--PQGMFFG 230
Query 178 SENISQAWTLGRIDCQISKGQ-CSSYVASYVN-----SSCTIPKVFKLSSVCPFNVHSQK 231
+ W G QI K + C Y+A Y + ++ PK F+ N S++
Sbjct 231 KVDQRGWWPHG--SSQIEKARNCVGYLAKYASKFTAITAAAFPKGFRTHGCGGLNTESKR 288
> xcc:XCC2058 gII; replication initiation protein
Length=346
Score = 41.2 bits (95), Expect = 0.44, Method: Compositional matrix adjust.
Identities = 32/120 (27%), Positives = 53/120 (44%), Gaps = 17/120 (14%)
Query 124 DLQLFFKRLRYYVSK------QCPSEKVRYFAVGEYGPVHFRPHYHILLFLQSDEALQVC 177
D+ FKR+R + ++ + E RY VGE FRPHYH++L++ + +
Sbjct 174 DVSELFKRMRGHFNRLKSGRARWNRESFRYVWVGEL-TQRFRPHYHVMLWV--PQGMFFG 230
Query 178 SENISQAWTLGRIDCQISKGQ-CSSYVASYVN-----SSCTIPKVFKLSSVCPFNVHSQK 231
+ W G QI K + C Y+A Y + ++ PK F+ N S++
Sbjct 231 KVDQRGWWPHG--SSQIEKARNCVGYLAKYASKFTAITAAAFPKGFRTHGCGGLNTESKR 288
> laa:WSI_05020 hypothetical protein
Length=405
Score = 40.8 bits (94), Expect = 0.54, Method: Compositional matrix adjust.
Identities = 47/171 (27%), Positives = 72/171 (42%), Gaps = 23/171 (13%)
Query 24 MVVPCGHCQACMLAKNSRYAFQCDLESYVAKHTLFITLT------YANRYIPRATFVDSL 77
+++PC C +C + + + +E + T F+TLT +AN ++DS
Sbjct 202 LILPCRSCSSCYKNRGLFWLRRAYIEVKRSTRTWFVTLTMTPANHFANHRSMVFNYIDSF 261
Query 78 ERPFGNDL--VDKETGEILGPSDMKQEDIDRLLNKFYLFGDVPYLRKTDLQLFFKRLRYY 135
P DL VD EI M+++DI + LF + + LF KRLR
Sbjct 262 P-PHERDLLNVDGRPTEI---HLMRKKDI---FGENVLFSLLCKGFGNKVSLFLKRLRKN 314
Query 136 VSKQCPSEKVRYFAVGEYGPVHFRPHYHILLFLQSDEALQVCSENISQAWT 186
K K RYF V E PH H+L+ Q D L+ + + W+
Sbjct 315 TGK-----KFRYFFVFEKHKSG-DPHVHMLIHQQCDNLLK--KAEVQEEWS 357
> plu:plu4620 recQ; ATP-dependent DNA helicase RecQ
Length=608
Score = 40.8 bits (94), Expect = 0.80, Method: Compositional matrix adjust.
Identities = 23/92 (25%), Positives = 41/92 (45%), Gaps = 0/92 (0%)
Query 398 RKSKQRMIEEFYSYLDYMHLTTFFESQQEFYESDLIGDDDLCTDQWENSYYPYFYYNVHT 457
R+ +Q MI+ Y + + + F E E+ + L D+ C QW + + P +
Sbjct 109 RRCRQGMIKLLYIAPERLMMDNFLEQLLEWQPAMLAVDEAHCISQWGHDFRPEYRALGQL 168
Query 458 NEPFEKTPVYRLYASDVKKLFNDRIKHKKLND 489
+ F PV L A+ + ND ++ LN+
Sbjct 169 RQRFPTLPVIALTATADETTRNDIVRLLNLNN 200
> pay:PAU_04111 recQ; ATP-dependent DNA helicase (EC:3.6.1.-)
Length=608
Score = 40.0 bits (92), Expect = 1.2, Method: Compositional matrix adjust.
Identities = 22/92 (24%), Positives = 41/92 (45%), Gaps = 0/92 (0%)
Query 398 RKSKQRMIEEFYSYLDYMHLTTFFESQQEFYESDLIGDDDLCTDQWENSYYPYFYYNVHT 457
R+ +Q M++ Y + + + F E E+ + L D+ C QW + + P +
Sbjct 109 RRCRQGMVKLLYIAPERLMMDNFLEQLLEWQPAILAVDEAHCISQWGHDFRPEYRALGQL 168
Query 458 NEPFEKTPVYRLYASDVKKLFNDRIKHKKLND 489
+ F PV L A+ + ND ++ LN+
Sbjct 169 RQRFPTLPVIALTATADETTRNDIVRLLNLNN 200
> rsd:TGRD_578 hypothetical protein
Length=175
Score = 38.1 bits (87), Expect = 2.0, Method: Compositional matrix adjust.
Identities = 25/62 (40%), Positives = 34/62 (55%), Gaps = 7/62 (11%)
Query 100 KQEDIDRLLNKFYLFGDVPYLRKTDLQLFFKRLRYYVSKQCPSEKVRYFAVGEYGPVHFR 159
+ ++ R LNK+ F RK D+Q F K LR Y+S + K+R F GEYG + R
Sbjct 117 EHNELVRELNKYSTF------RKLDVQTFIKDLRDYLSYH-RNLKIRCFYCGEYGNILDR 169
Query 160 PH 161
PH
Sbjct 170 PH 171
> top:TOPB45_0567 hypothetical protein
Length=674
Score = 38.1 bits (87), Expect = 5.4, Method: Compositional matrix adjust.
Identities = 45/169 (27%), Positives = 79/169 (47%), Gaps = 22/169 (13%)
Query 245 YSSTPQNFVKRSIVLNGKYKEFDVWRSCYAYYFPRCKGFASKSSRERAYSYGIYDTARRL 304
Y P N + I+ NG E ++ + + Y G ++ +E Y Y
Sbjct 140 YRDIPVNLIDLYILENG---ELNIIDNEFIKYLEEKSGM--ENLKELKYKLENY-----A 189
Query 305 FSSSETTFSLAKEIAFYIKHFHFTDDTYLLDLFGHVSDQKSLLDLSNYFLDR-DAM---- 359
FS S + E+ Y + F D+ L+D+ G V+ + D+S FLDR +A+
Sbjct 190 FSRSRDNIPIKIELG-YPFPWEF-DEIRLVDMPG-VNAVGGVHDISFSFLDRANAILFIH 246
Query 360 -VRPVESDEFNRWVHRIYTELLVSKHFLYFVCTHTTLAERKSKQRMIEE 407
++P+ES+ F R++ + T SK L+ + TH+ + + K R++EE
Sbjct 247 PIKPIESESFKRFISSVITN--KSKENLFLILTHSAVF-FEEKDRLLEE 292
> ear:ST548_p3284 FIG00947185: hypothetical protein
Length=194
Score = 36.6 bits (83), Expect = 8.2, Method: Compositional matrix adjust.
Identities = 28/80 (35%), Positives = 37/80 (46%), Gaps = 16/80 (20%)
Query 142 SEKVRYFAVGEYGPVHFRPHYHILLFLQSDEALQVCSEN--------ISQAW--TLGRID 191
S ++RY V E G ++ R HYH+LL L D V S N I QAW LG
Sbjct 77 SNRLRYAWVREVGELNRRKHYHVLLLLNKDFYHGVGSYNADDSLYALIQQAWCSALG--- 133
Query 192 CQISKGQCSSYVASYVNSSC 211
+ GQ S +A+ + C
Sbjct 134 --LDSGQYSG-LANMTENGC 150
Lambda K H a alpha
0.324 0.138 0.429 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 1084184891959