bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: all_orgs
14,240,465 sequences; 5,121,972,263 total letters
Query= Contig-14_CDS_annotation_glimmer3.pl_2_1
Length=346
Score E
Sequences producing significant alignments: (Bits) Value
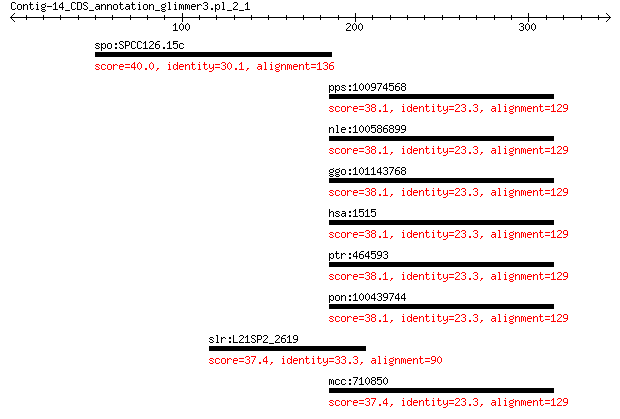
spo:SPCC126.15c sec65; signal recognition particle subunit Sec... 40.0 0.39
pps:100974568 CTSL2; cathepsin L2 38.1 2.0
nle:100586899 CTSL2; cathepsin L2 38.1 2.0
ggo:101143768 CTSL2; cathepsin L2 isoform 1 38.1
hsa:1515 CTSV, CATL2, CTSL2, CTSU; cathepsin V (EC:3.4.22.43) 38.1 2.0
ptr:464593 CTSL2; cathepsin L2 38.1 2.1
pon:100439744 CTSL2; cathepsin L2 38.1 2.1
slr:L21SP2_2619 hypothetical protein 37.4 3.3
mcc:710850 CTSL2; cathepsin L2 37.4 4.5
> spo:SPCC126.15c sec65; signal recognition particle subunit Sec65
(predicted)
Length=213
Score = 40.0 bits (92), Expect = 0.39, Method: Compositional matrix adjust.
Identities = 41/155 (26%), Positives = 63/155 (41%), Gaps = 30/155 (19%)
Query 50 ALKTYNSDLLQNWINT--------EWIEGEQGINEISAVDVSNGQLTMDALNLAQKVYNM 101
LKT+ +D W+N E I+ + INEI+ V + + D L+L + N+
Sbjct 65 PLKTHPAD----WVNPGRVEMVLPEKIQKKYVINEIAKVLLLRPTVKTDPLSLP--IQNV 118
Query 102 LNRIAVSGGTYRDWLETVFTGGNYMERCETPVFEGGMSQEIVFQEVISNS------ASGE 155
R+ + Y + GN + +P GG E +FQE++ A G
Sbjct 119 PARLPENPPAYPKGVL-----GNTILPLHSPALSGGGISENMFQEMMQEMQKQPGLAGGM 173
Query 156 EPLGTLAGRG-----ISTEKQKGGHVKIKVTEPCY 185
P+ LAG G + T + K K EP Y
Sbjct 174 NPMAALAGMGGPAPPMPTPQASSSQRKQKSIEPEY 208
> pps:100974568 CTSL2; cathepsin L2
Length=334
Score = 38.1 bits (87), Expect = 2.0, Method: Compositional matrix adjust.
Identities = 30/133 (23%), Positives = 51/133 (38%), Gaps = 20/133 (15%)
Query 185 YIIGIGSITPRIDYSQGNEFYAYHQTVDDIHKPALDGIGYQDSVNWQRAFWDRQYNT--- 241
+ +GI S P+ D + ++Y + T ++ +G W+RA W++
Sbjct 10 FCLGIASAVPKFDQNLDTKWYQWKATHRRLYGANEEG--------WRRAVWEKNMKMIEL 61
Query 242 -TGQIQQPAVGKTVAWINYMTNINRTFGNFADNNSEAFMVMNRNYEYRSGTTFGTTAIND 300
G+ Q G T+A FG+ + M RN ++R G F D
Sbjct 62 HNGEYSQGKHGFTMA--------MNAFGDMTNEEFRQMMGCFRNQKFRKGKVFREPLFLD 113
Query 301 LTTYIDPVKFNYI 313
L +D K Y+
Sbjct 114 LPKSVDWRKKGYV 126
> nle:100586899 CTSL2; cathepsin L2
Length=334
Score = 38.1 bits (87), Expect = 2.0, Method: Compositional matrix adjust.
Identities = 30/133 (23%), Positives = 51/133 (38%), Gaps = 20/133 (15%)
Query 185 YIIGIGSITPRIDYSQGNEFYAYHQTVDDIHKPALDGIGYQDSVNWQRAFWDRQYNT--- 241
+ +GI S P+ D + ++Y + T ++ +G W+RA W++
Sbjct 10 FCLGIASAVPKFDQNLDTKWYQWKATHRRLYGANEEG--------WRRAVWEKNMKMIEL 61
Query 242 -TGQIQQPAVGKTVAWINYMTNINRTFGNFADNNSEAFMVMNRNYEYRSGTTFGTTAIND 300
G+ Q G T+A FG+ + M RN ++R G F D
Sbjct 62 HNGEYSQGKHGFTMA--------MNAFGDMTNEEFRQMMGCFRNQKFRKGKVFREPLFLD 113
Query 301 LTTYIDPVKFNYI 313
L +D K Y+
Sbjct 114 LPKSVDWRKKGYV 126
> ggo:101143768 CTSL2; cathepsin L2 isoform 1
Length=334
Score = 38.1 bits (87), Expect = 2.0, Method: Compositional matrix adjust.
Identities = 30/133 (23%), Positives = 51/133 (38%), Gaps = 20/133 (15%)
Query 185 YIIGIGSITPRIDYSQGNEFYAYHQTVDDIHKPALDGIGYQDSVNWQRAFWDRQYNT--- 241
+ +GI S P+ D + ++Y + T ++ +G W+RA W++
Sbjct 10 FCLGIASAVPKFDQNLDTKWYQWKATHRRLYGANEEG--------WRRAVWEKNMKMIEL 61
Query 242 -TGQIQQPAVGKTVAWINYMTNINRTFGNFADNNSEAFMVMNRNYEYRSGTTFGTTAIND 300
G+ Q G T+A FG+ + M RN ++R G F D
Sbjct 62 HNGEYSQGKHGFTMA--------MNAFGDMTNEEFRQMMGCFRNQKFRKGKVFREPLFLD 113
Query 301 LTTYIDPVKFNYI 313
L +D K Y+
Sbjct 114 LPKSVDWRKKGYV 126
> hsa:1515 CTSV, CATL2, CTSL2, CTSU; cathepsin V (EC:3.4.22.43)
Length=334
Score = 38.1 bits (87), Expect = 2.0, Method: Compositional matrix adjust.
Identities = 30/133 (23%), Positives = 51/133 (38%), Gaps = 20/133 (15%)
Query 185 YIIGIGSITPRIDYSQGNEFYAYHQTVDDIHKPALDGIGYQDSVNWQRAFWDRQYNT--- 241
+ +GI S P+ D + ++Y + T ++ +G W+RA W++
Sbjct 10 FCLGIASAVPKFDQNLDTKWYQWKATHRRLYGANEEG--------WRRAVWEKNMKMIEL 61
Query 242 -TGQIQQPAVGKTVAWINYMTNINRTFGNFADNNSEAFMVMNRNYEYRSGTTFGTTAIND 300
G+ Q G T+A FG+ + M RN ++R G F D
Sbjct 62 HNGEYSQGKHGFTMA--------MNAFGDMTNEEFRQMMGCFRNQKFRKGKVFREPLFLD 113
Query 301 LTTYIDPVKFNYI 313
L +D K Y+
Sbjct 114 LPKSVDWRKKGYV 126
> ptr:464593 CTSL2; cathepsin L2
Length=334
Score = 38.1 bits (87), Expect = 2.1, Method: Compositional matrix adjust.
Identities = 30/133 (23%), Positives = 51/133 (38%), Gaps = 20/133 (15%)
Query 185 YIIGIGSITPRIDYSQGNEFYAYHQTVDDIHKPALDGIGYQDSVNWQRAFWDRQYNT--- 241
+ +GI S P+ D + ++Y + T ++ +G W+RA W++
Sbjct 10 FCLGIASAVPKFDQNLDTKWYQWKATHRRLYGANEEG--------WRRAVWEKNMKMIEL 61
Query 242 -TGQIQQPAVGKTVAWINYMTNINRTFGNFADNNSEAFMVMNRNYEYRSGTTFGTTAIND 300
G+ Q G T+A FG+ + M RN ++R G F D
Sbjct 62 HNGEYSQGKHGFTMA--------MNAFGDMTNEEFRQMMGCFRNQKFRKGKVFREPLFLD 113
Query 301 LTTYIDPVKFNYI 313
L +D K Y+
Sbjct 114 LPKSVDWRKKGYV 126
> pon:100439744 CTSL2; cathepsin L2
Length=334
Score = 38.1 bits (87), Expect = 2.1, Method: Compositional matrix adjust.
Identities = 30/133 (23%), Positives = 51/133 (38%), Gaps = 20/133 (15%)
Query 185 YIIGIGSITPRIDYSQGNEFYAYHQTVDDIHKPALDGIGYQDSVNWQRAFWDRQYNT--- 241
+ +GI S P+ D + ++Y + T ++ +G W+RA W++
Sbjct 10 FCLGIASAVPKFDQNLDTKWYQWKATHRRLYGANEEG--------WRRAVWEKNMKMIEL 61
Query 242 -TGQIQQPAVGKTVAWINYMTNINRTFGNFADNNSEAFMVMNRNYEYRSGTTFGTTAIND 300
G+ Q G T+A FG+ + M RN ++R G F D
Sbjct 62 HNGEYSQGKHGFTMA--------MNAFGDMTNEEFRQMMGCFRNQKFRKGKVFREPLFLD 113
Query 301 LTTYIDPVKFNYI 313
L +D K Y+
Sbjct 114 LPKSVDWRKKGYV 126
> slr:L21SP2_2619 hypothetical protein
Length=271
Score = 37.4 bits (85), Expect = 3.3, Method: Compositional matrix adjust.
Identities = 30/101 (30%), Positives = 44/101 (44%), Gaps = 16/101 (16%)
Query 116 LETVFTGGNYMERCE-----------TPVFEGGMSQEIVFQEVISNSASGEEPLGTLAGR 164
LET GGNY R + P G + I ++ + S +G LAGR
Sbjct 58 LETANLGGNYSGRSKPEILQTDGEVSAPALTGAGNGTISLHGSLTMARS----MGFLAGR 113
Query 165 GISTEKQKGGHVKIKVTEPCYIIGIGSITPRIDYSQGNEFY 205
S E + G + + E +I G G++ R+D +G EFY
Sbjct 114 IASRETAQPGDIFAALVEDHFIPGPGAVVHRVDLPEG-EFY 153
> mcc:710850 CTSL2; cathepsin L2
Length=334
Score = 37.4 bits (85), Expect = 4.5, Method: Compositional matrix adjust.
Identities = 30/133 (23%), Positives = 51/133 (38%), Gaps = 20/133 (15%)
Query 185 YIIGIGSITPRIDYSQGNEFYAYHQTVDDIHKPALDGIGYQDSVNWQRAFWDRQYNT--- 241
+ +GI S P+ D + ++Y + T ++ + +G W+RA W++
Sbjct 10 FCLGIASAVPKFDQNLDTKWYQWKATHRRLYGASEEG--------WRRAVWEKNMKMIEL 61
Query 242 -TGQIQQPAVGKTVAWINYMTNINRTFGNFADNNSEAFMVMNRNYEYRSGTTFGTTAIND 300
G+ Q G T+A FG+ + M RN + R G F D
Sbjct 62 HNGEYSQGKHGFTMA--------MNAFGDMTNEEFRQVMGCFRNQKLRKGKLFREPLFLD 113
Query 301 LTTYIDPVKFNYI 313
L +D K Y+
Sbjct 114 LPKSVDWRKKGYV 126
Lambda K H a alpha
0.317 0.133 0.397 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 644431275578