bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: all_orgs
14,240,465 sequences; 5,121,972,263 total letters
Query= Contig-21_CDS_annotation_glimmer3.pl_2_1
Length=107
Score E
Sequences producing significant alignments: (Bits) Value
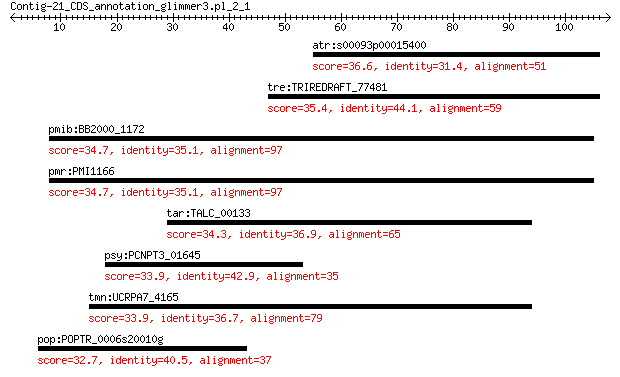
atr:s00093p00015400 AMTR_s00093p00015400; hypothetical protein 36.6 0.55
tre:TRIREDRAFT_77481 hypothetical protein 35.4 1.2
pmib:BB2000_1172 fadD; long-chain-fatty-acid--CoA ligase 34.7 2.1
pmr:PMI1166 fadD; long-chain-fatty-acid--CoA ligase (EC:6.2.1.3) 34.7 2.1
tar:TALC_00133 DNA modification methylase (EC:2.1.1.72) 34.3 3.2
psy:PCNPT3_01645 phosphatidylserine decarboxylase 33.9 3.5
tmn:UCRPA7_4165 putative wd repeat-containing protein 2 protein 33.9 4.1
pop:POPTR_0006s20010g hypothetical protein 32.7 9.6
> atr:s00093p00015400 AMTR_s00093p00015400; hypothetical protein
Length=589
Score = 36.6 bits (83), Expect = 0.55, Method: Composition-based stats.
Identities = 16/53 (30%), Positives = 32/53 (60%), Gaps = 2/53 (4%)
Query 55 LPEFDIRT--DRFEIAIDAMDKINQSAASQIAKSSGETEAVKDFGTETKTDPE 105
+ EFD + + FE+A +++ + +I + +GETE ++++G +TDPE
Sbjct 5 MAEFDCESVIEAFEVATKDAERVQRETLRRILEENGETEYLQEWGLRGRTDPE 57
> tre:TRIREDRAFT_77481 hypothetical protein
Length=812
Score = 35.4 bits (80), Expect = 1.2, Method: Composition-based stats.
Identities = 26/66 (39%), Positives = 38/66 (58%), Gaps = 8/66 (12%)
Query 47 YTEKKDGVLP-EFDIR--TDRFEIAIDAMDKI----NQSAASQIAKSSGETEAVKDFGTE 99
Y EK + P E IR TDRF +AIDA+D++ N+ AA++ A + + EA ++ E
Sbjct 731 YKEKGNIDTPLELAIRNQTDRFSLAIDAIDRMPHLHNKGAAARQAMLNAQIEA-RNEAFE 789
Query 100 TKTDPE 105
DPE
Sbjct 790 KGMDPE 795
> pmib:BB2000_1172 fadD; long-chain-fatty-acid--CoA ligase
Length=562
Score = 34.7 bits (78), Expect = 2.1, Method: Composition-based stats.
Identities = 34/99 (34%), Positives = 51/99 (52%), Gaps = 10/99 (10%)
Query 8 KGCINNPDLTYQAEPREVKLRKIISGESSSMEDGVFPTIYTEKKDGVLPE-FDIRTDRFE 66
KG N PD T + + + +G+ ++M+D F I KKD +L F++ + E
Sbjct 416 KGYWNRPDATAEV----LHDGWVATGDIATMDDEGFIRIIDRKKDMILVSGFNVYPNEVE 471
Query 67 IAIDAMDKINQSAASQI-AKSSGETEAVKDFGTETKTDP 104
+ A K+ +SAA + +KSSGET VK F K DP
Sbjct 472 EVVTAHPKVLESAAIGVPSKSSGET--VKIF--VVKKDP 506
> pmr:PMI1166 fadD; long-chain-fatty-acid--CoA ligase (EC:6.2.1.3)
Length=562
Score = 34.7 bits (78), Expect = 2.1, Method: Composition-based stats.
Identities = 34/99 (34%), Positives = 51/99 (52%), Gaps = 10/99 (10%)
Query 8 KGCINNPDLTYQAEPREVKLRKIISGESSSMEDGVFPTIYTEKKDGVLPE-FDIRTDRFE 66
KG N PD T + + + +G+ ++M+D F I KKD +L F++ + E
Sbjct 416 KGYWNRPDATAEV----LHDGWVATGDIATMDDEGFIRIIDRKKDMILVSGFNVYPNEVE 471
Query 67 IAIDAMDKINQSAASQI-AKSSGETEAVKDFGTETKTDP 104
+ A K+ +SAA + +KSSGET VK F K DP
Sbjct 472 EVVTAHPKVLESAAIGVPSKSSGET--VKIF--VVKKDP 506
> tar:TALC_00133 DNA modification methylase (EC:2.1.1.72)
Length=379
Score = 34.3 bits (77), Expect = 3.2, Method: Compositional matrix adjust.
Identities = 24/65 (37%), Positives = 36/65 (55%), Gaps = 3/65 (5%)
Query 29 KIISGESSSMEDGVFPTIYTEKKDGVLPEFDIRTDRFEIAIDAMDKINQSAASQIAKSSG 88
KI ++S + FP I+T+ K G LPE++ RTD + ++I M K N +I SG
Sbjct 9 KIAREIANSWLEEYFPEIHTKLKIG-LPEYNDRTDNWHVSI--MIKNNSENIGEIRIDSG 65
Query 89 ETEAV 93
T+ V
Sbjct 66 LTKVV 70
> psy:PCNPT3_01645 phosphatidylserine decarboxylase
Length=300
Score = 33.9 bits (76), Expect = 3.5, Method: Compositional matrix adjust.
Identities = 15/38 (39%), Positives = 24/38 (63%), Gaps = 3/38 (8%)
Query 18 YQAEPREVKLRKIISGE---SSSMEDGVFPTIYTEKKD 52
+QA+ + LR+++ G+ S+ EDG+F TIY KD
Sbjct 123 FQAKGHDFSLRELLGGQDAISAPFEDGIFSTIYLAPKD 160
> tmn:UCRPA7_4165 putative wd repeat-containing protein 2 protein
Length=605
Score = 33.9 bits (76), Expect = 4.1, Method: Composition-based stats.
Identities = 29/87 (33%), Positives = 44/87 (51%), Gaps = 14/87 (16%)
Query 15 DLTYQAE--PREVKLRKIISGESSSM------EDGVFPTIYTEKKDGVLPEFDIRTDRFE 66
DL Y AE P+ +K ++ G + S+ DG T+YT DG++ +++I T
Sbjct 306 DLNYLAEGSPKPIK---VVQGHNKSITAIGAASDGKGETVYTGSFDGLVNQWNIGTG-LA 361
Query 67 IAIDAMDKINQSAASQIAKSSGETEAV 93
A+D NQ +Q A SSG T +V
Sbjct 362 TAVDGQTHTNQ--VTQFATSSGRTYSV 386
> pop:POPTR_0006s20010g hypothetical protein
Length=365
Score = 32.7 bits (73), Expect = 9.6, Method: Compositional matrix adjust.
Identities = 15/37 (41%), Positives = 25/37 (68%), Gaps = 0/37 (0%)
Query 6 NRKGCINNPDLTYQAEPREVKLRKIISGESSSMEDGV 42
NRKG I+ + +A+PR +K+RK SGE +++D +
Sbjct 50 NRKGTIDPALMAAEAKPRTIKVRKDGSGEFKTLKDAI 86
Lambda K H a alpha
0.309 0.128 0.344 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 125230604613