bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: all_orgs
14,240,465 sequences; 5,121,972,263 total letters
Query= Contig-21_CDS_annotation_glimmer3.pl_2_3
Length=101
Score E
Sequences producing significant alignments: (Bits) Value
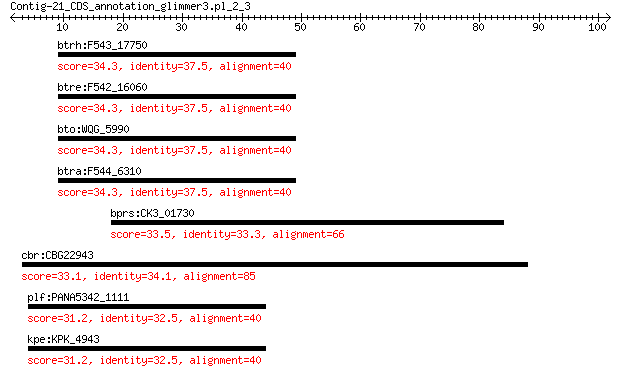
btrh:F543_17750 3'(2'),5'-bisphosphate nucleotidase CysQ 34.3 2.3
btre:F542_16060 3'(2'),5'-bisphosphate nucleotidase CysQ 34.3 2.3
bto:WQG_5990 3'(2'),5'-bisphosphate nucleotidase CysQ 34.3 2.3
btra:F544_6310 3'(2'),5'-bisphosphate nucleotidase CysQ 34.3 2.4
bprs:CK3_01730 CoA-substrate-specific enzyme activase, putative 33.5 4.9
cbr:CBG22943 Cbr-aco-2; C. briggsae CBR-ACO-2 protein 33.1 6.3
plf:PANA5342_1111 hypothetical protein 31.2 7.5
kpe:KPK_4943 hypothetical protein 31.2 7.8
> btrh:F543_17750 3'(2'),5'-bisphosphate nucleotidase CysQ
Length=272
Score = 34.3 bits (77), Expect = 2.3, Method: Compositional matrix adjust.
Identities = 15/40 (38%), Positives = 23/40 (58%), Gaps = 0/40 (0%)
Query 9 LLAESGKEEEEIKDVNLEIEEREVSENGPFVLIRNKKNKW 48
LLAE G E ++ + L +RE N PF+++ NK+ W
Sbjct 224 LLAECGGEIFDLNFLPLTYNQRETFINPPFIMVGNKQYNW 263
> btre:F542_16060 3'(2'),5'-bisphosphate nucleotidase CysQ
Length=272
Score = 34.3 bits (77), Expect = 2.3, Method: Compositional matrix adjust.
Identities = 15/40 (38%), Positives = 23/40 (58%), Gaps = 0/40 (0%)
Query 9 LLAESGKEEEEIKDVNLEIEEREVSENGPFVLIRNKKNKW 48
LLAE G E ++ + L +RE N PF+++ NK+ W
Sbjct 224 LLAECGGEIFDLNFLPLTYNQRETFINPPFIMVGNKQYNW 263
> bto:WQG_5990 3'(2'),5'-bisphosphate nucleotidase CysQ
Length=272
Score = 34.3 bits (77), Expect = 2.3, Method: Compositional matrix adjust.
Identities = 15/40 (38%), Positives = 23/40 (58%), Gaps = 0/40 (0%)
Query 9 LLAESGKEEEEIKDVNLEIEEREVSENGPFVLIRNKKNKW 48
LLAE G E ++ + L +RE N PF+++ NK+ W
Sbjct 224 LLAECGGEIFDLNFLPLTYNQRETFINPPFIMVGNKQYNW 263
> btra:F544_6310 3'(2'),5'-bisphosphate nucleotidase CysQ
Length=272
Score = 34.3 bits (77), Expect = 2.4, Method: Compositional matrix adjust.
Identities = 15/40 (38%), Positives = 23/40 (58%), Gaps = 0/40 (0%)
Query 9 LLAESGKEEEEIKDVNLEIEEREVSENGPFVLIRNKKNKW 48
LLAE G E ++ + L +RE N PF+++ NK+ W
Sbjct 224 LLAECGGEIFDLNFLPLTYNQRETFINPPFIMVGNKQYNW 263
> bprs:CK3_01730 CoA-substrate-specific enzyme activase, putative
Length=1141
Score = 33.5 bits (75), Expect = 4.9, Method: Composition-based stats.
Identities = 22/66 (33%), Positives = 33/66 (50%), Gaps = 3/66 (5%)
Query 18 EEIKDVNLEIEEREVSENGPFVLIRNKKNKWVITTCGALVNGKEFDTKEDAEKHLAKKEW 77
E IK+ E++E+ V PF+ ++ V++ V KEFD E K AKK W
Sbjct 773 ENIKNNMEELKEKNVKFLNPFMAFTSEA---VLSKQLQEVFKKEFDIPESETKKAAKKAW 829
Query 78 DDILTA 83
D++ A
Sbjct 830 DELAQA 835
> cbr:CBG22943 Cbr-aco-2; C. briggsae CBR-ACO-2 protein
Length=777
Score = 33.1 bits (74), Expect = 6.3, Method: Composition-based stats.
Identities = 29/97 (30%), Positives = 45/97 (46%), Gaps = 14/97 (14%)
Query 3 KKFRDQLLAESGKEEEEIKDVNLEIEEREVSENGPFV---------LIRN-KKNKWVITT 52
+K++D L A+ G ++I ++NL+ V NGPF L N KKN W +
Sbjct 317 RKYKDLLTADDGAHYDQIIEINLDTLTPHV--NGPFTPDLASPIDKLGENAKKNDWPLDV 374
Query 53 CGALVNGKEFDTKEDAEK--HLAKKEWDDILTAALIF 87
+L+ + ED + +AK+ D L A IF
Sbjct 375 KVSLIGSCTNSSYEDMTRAASIAKQALDKGLKAKTIF 411
> plf:PANA5342_1111 hypothetical protein
Length=73
Score = 31.2 bits (69), Expect = 7.5, Method: Compositional matrix adjust.
Identities = 13/40 (33%), Positives = 23/40 (58%), Gaps = 0/40 (0%)
Query 4 KFRDQLLAESGKEEEEIKDVNLEIEEREVSENGPFVLIRN 43
K+ + +G+E E+I+ V ER NGP+V++R+
Sbjct 30 KWSGSICHYAGREVEDIRGVLAVFAERRQDRNGPYVILRS 69
> kpe:KPK_4943 hypothetical protein
Length=73
Score = 31.2 bits (69), Expect = 7.8, Method: Compositional matrix adjust.
Identities = 13/40 (33%), Positives = 23/40 (58%), Gaps = 0/40 (0%)
Query 4 KFRDQLLAESGKEEEEIKDVNLEIEEREVSENGPFVLIRN 43
K+ + +G+E E+I+ V ER NGP+V++R+
Sbjct 30 KWSGSICHYAGREVEDIRGVLAVFAERRQDRNGPYVILRS 69
Lambda K H a alpha
0.311 0.130 0.361 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 127879331103