bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: all_orgs
14,240,465 sequences; 5,121,972,263 total letters
Query= Contig-25_CDS_annotation_glimmer3.pl_2_1
Length=449
Score E
Sequences producing significant alignments: (Bits) Value
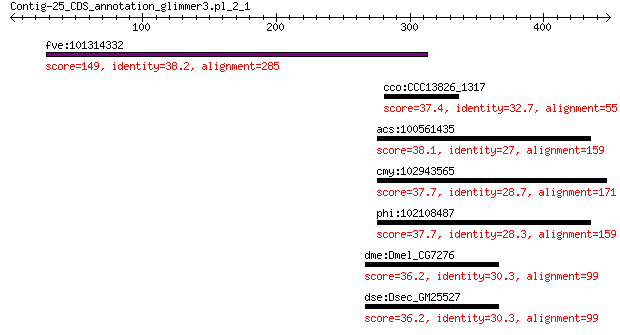
fve:101314332 capsid protein VP1-like 149 2e-37
cco:CCC13826_1317 2-hydroxyglutaryl-CoA dehydratase, D-component 37.4 3.7
acs:100561435 jph1; junctophilin 1 38.1
cmy:102943565 JPH1; junctophilin 1 37.7
phi:102108487 JPH1; junctophilin 1 37.7
dme:Dmel_CG7276 CG7276 gene product from transcript CG7276-RB ... 36.2 6.9
dse:Dsec_GM25527 GM25527 gene product from transcript GM25527-RA 36.2 7.7
> fve:101314332 capsid protein VP1-like
Length=421
Score = 149 bits (377), Expect = 2e-37, Method: Compositional matrix adjust.
Identities = 109/298 (37%), Positives = 154/298 (52%), Gaps = 42/298 (14%)
Query 28 GSIADYLGLPI-GSISQSSPVS--VLPFRCFALIYDKYFRNENTTDEIYIQKKGFSLSEL 84
GS+ DY GLP G + S+ VS LP R + LIY+++FR+EN + + + K
Sbjct 153 GSLQDYFGLPTAGQVGVSNTVSHSALPVRAYNLIYNQWFRDENLQNSVVVDKGD------ 206
Query 85 IGAQNFSPNSYCGKLPKVNKYKDYFTSCVPNPQKGA-PVTFNLGDQAVV-----RTSDSE 138
G +Y L + K DYFTS +P PQKG V+ LG A + SD
Sbjct 207 -GPDTTPSTNYT--LLRRGKRHDYFTSALPWPQKGGTAVSLPLGTSAPIAFSGASGSDVG 263
Query 139 LVTGPQEQMALTNSQSGSASVGEHPLIVGLGGMRFDAAAFSGTVAAGLYPNNLYADLSSA 198
+++ Q + +GS + L +G S TVA GLY ADLS+A
Sbjct 264 VISTTQGNLIKNMYSTGSGT----SLKIG-----------SATVATGLY-----ADLSAA 303
Query 199 NAISVDDLRLAFAYQKMLERDAIYGSRYNEYLYGHFGVHIPDAYIQFPQYLGGGRTPLNI 258
A +++ LR +F QK+LERDA G+RY E + HFGV PDA +Q P+YLGGG TP+NI
Sbjct 304 TAATINQLRQSFQIQKLLERDARGGTRYTEIIRSHFGVASPDARLQRPEYLGGGSTPINI 363
Query 259 VQV---AQTSQGTEESPLGNVGAY-SWTNGRTGYSRKFKEHGIVMTVACLRYRHTYQQ 312
+ T +P GN+ A+ ++ G+S+ F EHG V+ + +R TYQQ
Sbjct 364 APIAQTGGTGAQGTTTPQGNLAAFGTYMAKGHGFSQSFVEHGHVIGLVSVRADLTYQQ 421
> cco:CCC13826_1317 2-hydroxyglutaryl-CoA dehydratase, D-component
Length=183
Score = 37.4 bits (85), Expect = 3.7, Method: Compositional matrix adjust.
Identities = 18/55 (33%), Positives = 26/55 (47%), Gaps = 7/55 (13%)
Query 281 WTNGRTGYSRKFKEHGIVMTVACLRYRHTYQQGIAKKWRRKVREDFYDPLFSTIG 335
W+N G ++ K + C R+ H Q +KKW RK+ D+ DP IG
Sbjct 83 WSNYVDGCLKEEK-------IYCERFSHIVQNQFSKKWFRKIFNDYQDPSIKAIG 130
> acs:100561435 jph1; junctophilin 1
Length=654
Score = 38.1 bits (87), Expect = 4.7, Method: Compositional matrix adjust.
Identities = 43/171 (25%), Positives = 71/171 (42%), Gaps = 29/171 (17%)
Query 276 VGAYSWTNGRT--GYSRKFKEHGI-VMTVACLRYRHTYQQGIAKKWRRKVREDFYDPL-- 330
VGAY+W +G T G+ + K HG+ V + YR + G K R VR+ +P
Sbjct 49 VGAYTWPSGNTYQGHWAQGKRHGLGVESKGKWTYRGEWAHGF--KGRYGVRQSLANPARY 106
Query 331 --FSTIGQQPVYTTELYAQAFP---QTVFGYREAWSELRNIPNTISGEMRSGVTNSLDIW 385
+ G Q Y E Y Q G R + +++P ++ +RS + SL
Sbjct 107 EGTWSNGLQDGYGVETYGDGGTYQGQWTGGMRHGYGMRQSVPYGMATVIRSPLRTSL--- 163
Query 386 HFADNYSSSPTLSQSFTEETPSYV--DRTLSVPSSSQNNFILNFYFDMSAV 434
+ +E++ V D T P+ ++ F+LNF D+ +
Sbjct 164 ------------ASLRSEQSNGTVLHDITSDSPAGTRGGFVLNFQSDLEGI 202
> cmy:102943565 JPH1; junctophilin 1
Length=626
Score = 37.7 bits (86), Expect = 5.0, Method: Compositional matrix adjust.
Identities = 49/187 (26%), Positives = 80/187 (43%), Gaps = 34/187 (18%)
Query 276 VGAYSWTNGRT--GYSRKFKEHGI-VMTVACLRYRHTYQQGIAKKWRRKVREDF-----Y 327
VG Y+W +G T GY + K HG+ V T YR + G K R VR+ Y
Sbjct 12 VGGYTWPSGNTYQGYWAQGKRHGLGVETKGRWMYRGEWSHGF--KGRYGVRQSLSTPARY 69
Query 328 DPLFSTIGQQPVYTTELYAQAFP---QTVFGYREAWSELRNIPNTISGEMRSGVTNSLDI 384
+ +S G Q Y E Y Q G R + +++P ++ +RS + SL
Sbjct 70 EGTWSN-GLQDGYGVETYGDGGTYQGQWTGGMRHGYGVRQSVPYGMATVIRSPLRTSL-- 126
Query 385 WHFADNYSSSPTLSQSFTEETPSYV--DRTLSVPSSSQNNFILNFYFD---MSAVRKMPV 439
+ +E++ V D T P+ ++ F+LNF+ D MS+ ++ +
Sbjct 127 -------------ASLRSEQSNGTVLHDITSDSPAGTRGGFVLNFHSDPEAMSSKKRGGL 173
Query 440 YSMPSLI 446
+ SL+
Sbjct 174 FRRGSLL 180
> phi:102108487 JPH1; junctophilin 1
Length=665
Score = 37.7 bits (86), Expect = 6.1, Method: Compositional matrix adjust.
Identities = 45/172 (26%), Positives = 71/172 (41%), Gaps = 31/172 (18%)
Query 276 VGAYSWTNGRT--GYSRKFKEHGI-VMTVACLRYRHTYQQGIAKKWRRKVREDF-----Y 327
G Y+W +G T GY + K HG+ V T YR + G K R VR+ Y
Sbjct 49 AGGYTWPSGNTYLGYWAQGKRHGLGVETKGRWMYRGEWSHGF--KGRYGVRQSLSTPARY 106
Query 328 DPLFSTIGQQPVYTTELYAQAFP---QTVFGYREAWSELRNIPNTISGEMRSGVTNSLDI 384
+ +S G Q Y E Y Q G R + +++P ++ +RS + SL
Sbjct 107 EGTWSN-GLQDGYGVETYGDGGTYQGQWTGGMRHGYGVRQSVPYGMATVIRSPLRTSL-- 163
Query 385 WHFADNYSSSPTLSQSFTEETPSYV--DRTLSVPSSSQNNFILNFYFDMSAV 434
+ +E++ V D T P+ ++ F+LNF+ D A+
Sbjct 164 -------------ASLRSEQSNGTVLHDVTSDSPAGTRGGFVLNFHSDAEAM 202
> dme:Dmel_CG7276 CG7276 gene product from transcript CG7276-RB
(EC:3.6.1.3)
Length=171
Score = 36.2 bits (82), Expect = 6.9, Method: Compositional matrix adjust.
Identities = 30/107 (28%), Positives = 44/107 (41%), Gaps = 9/107 (8%)
Query 267 GTEESPLGNVGAYSWTNGRTGYSRKFKEHGIVMTVACLRYRH----TYQQGIAKKWRRKV 322
G E+ P +V Y++ N + +KF I + +A + TY Q A KW R+V
Sbjct 42 GNEDKPASDVTIYNYYNMGPAFGKKFPLPYIRLMIAQVVKEKLANKTYNQSEALKWTREV 101
Query 323 REDFYDPLFSTIGQQP----VYTTELYAQAFPQTVFGYREAWSELRN 365
+D + G P V LY Q +G R W EL +
Sbjct 102 ADDI-NIKMKGRGSSPRFKHVVNVMLYQQTGAGCFYGARAIWDELSD 147
> dse:Dsec_GM25527 GM25527 gene product from transcript GM25527-RA
Length=171
Score = 36.2 bits (82), Expect = 7.7, Method: Compositional matrix adjust.
Identities = 30/107 (28%), Positives = 44/107 (41%), Gaps = 9/107 (8%)
Query 267 GTEESPLGNVGAYSWTNGRTGYSRKFKEHGIVMTVACLRYRH----TYQQGIAKKWRRKV 322
G E+ P +V Y++ N + +KF I + +A + TY Q A KW R+V
Sbjct 42 GNEDKPASDVTIYNYYNMGPAFGKKFPLPYIRLMIAQVVKEKLANKTYNQSEALKWTREV 101
Query 323 REDFYDPLFSTIGQQP----VYTTELYAQAFPQTVFGYREAWSELRN 365
+D + G P V LY Q +G R W EL +
Sbjct 102 ADDI-NIKMKGRGSSPRFKHVVNVMLYQQTGAGCFYGARAIWDELSD 147
Lambda K H a alpha
0.317 0.134 0.400 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 944189245008