bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: all_orgs
14,240,465 sequences; 5,121,972,263 total letters
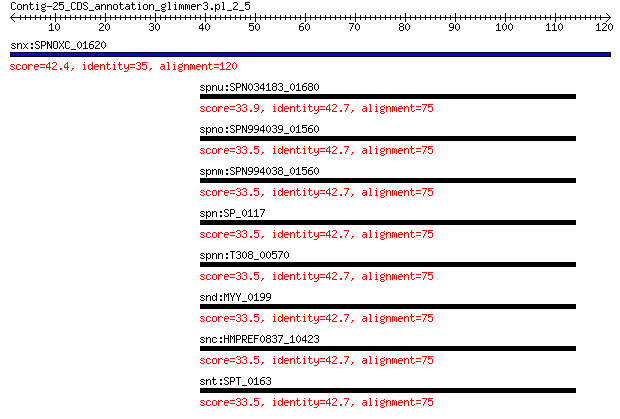
Query= Contig-25_CDS_annotation_glimmer3.pl_2_5
Length=120
Score E
Sequences producing significant alignments: (Bits) Value
snx:SPNOXC_01620 pspA; pneumococcal surface protein PspA 42.4 0.009
spnu:SPN034183_01680 pspA; pneumococcal surface protein PspA 33.9 7.0
spno:SPN994039_01560 pspA; pneumococcal surface protein PspA 33.5 7.4
spnm:SPN994038_01560 pspA; pneumococcal surface protein PspA 33.5 7.4
spn:SP_0117 surface protein A 33.5 8.8
spnn:T308_00570 choline binding protein J 33.5 9.2
snd:MYY_0199 surface protein A 33.5 9.2
snc:HMPREF0837_10423 psaA; manganese ABC transporter substrate... 33.5 9.2
snt:SPT_0163 pneumococcal surface protein A 33.5 9.2
> snx:SPNOXC_01620 pspA; pneumococcal surface protein PspA
Length=848
Score = 42.4 bits (98), Expect = 0.009, Method: Composition-based stats.
Identities = 42/142 (30%), Positives = 60/142 (42%), Gaps = 24/142 (17%)
Query 1 LQSCLERFIPQEDTTDEIQDNYDEYLDDLDGFTEYLDK--ADMYREKFNLSDELSAEEIF 58
L+ L+ P+ T DE+ E +LD E L AD+ +E NL L +
Sbjct 353 LEKLLDSLDPEGKTQDELDKEAAEA--ELDKKVESLQNKVADLEKEISNLEILLGGADSE 410
Query 59 DHVSEQAKKLKTRLEEIEKARKESQAKKHEPVPD-------------------KSTEKPQ 99
D + KL T+ E+EK +KE A +E PD E+P
Sbjct 411 DDTAALQNKLATKKAELEKTQKELDAALNELGPDGDEEETPAPAPQPEQPAPAPKPEQPA 470
Query 100 PQPQP-QPQPQPEPKKIEKEVK 120
P P+P QP P P+P++ K K
Sbjct 471 PAPKPEQPAPAPKPEQPAKPEK 492
> spnu:SPN034183_01680 pspA; pneumococcal surface protein PspA
Length=813
Score = 33.9 bits (76), Expect = 7.0, Method: Compositional matrix adjust.
Identities = 32/93 (34%), Positives = 41/93 (44%), Gaps = 18/93 (19%)
Query 39 ADMYREKFNLSDELSAEEIFDHVSEQAKKLKTRLEEIEKARKESQAKKHEPVPDKSTE-- 96
AD+ +E NL L + D + KL T+ E+EK +KE A +E PD E
Sbjct 388 ADLEKEISNLEILLGGADSEDDTAALQNKLATKKAELEKTQKELDAALNELGPDGDEEET 447
Query 97 --------------KP-QPQPQPQP-QPQPEPK 113
KP QP P P+P QP P PK
Sbjct 448 PAPAPQPEQPAPAPKPEQPAPAPKPEQPAPAPK 480
> spno:SPN994039_01560 pspA; pneumococcal surface protein PspA
Length=767
Score = 33.5 bits (75), Expect = 7.4, Method: Compositional matrix adjust.
Identities = 32/93 (34%), Positives = 41/93 (44%), Gaps = 18/93 (19%)
Query 39 ADMYREKFNLSDELSAEEIFDHVSEQAKKLKTRLEEIEKARKESQAKKHEPVPDKSTE-- 96
AD+ +E NL L + D + KL T+ E+EK +KE A +E PD E
Sbjct 391 ADLEKEISNLEILLGGADSEDDTAALQNKLATKKAELEKTQKELDAALNELGPDGDEEET 450
Query 97 --------------KP-QPQPQPQP-QPQPEPK 113
KP QP P P+P QP P PK
Sbjct 451 PAPAPQPEQPAPAPKPEQPAPAPKPEQPAPAPK 483
> spnm:SPN994038_01560 pspA; pneumococcal surface protein PspA
Length=767
Score = 33.5 bits (75), Expect = 7.4, Method: Compositional matrix adjust.
Identities = 32/93 (34%), Positives = 41/93 (44%), Gaps = 18/93 (19%)
Query 39 ADMYREKFNLSDELSAEEIFDHVSEQAKKLKTRLEEIEKARKESQAKKHEPVPDKSTE-- 96
AD+ +E NL L + D + KL T+ E+EK +KE A +E PD E
Sbjct 391 ADLEKEISNLEILLGGADSEDDTAALQNKLATKKAELEKTQKELDAALNELGPDGDEEET 450
Query 97 --------------KP-QPQPQPQP-QPQPEPK 113
KP QP P P+P QP P PK
Sbjct 451 PAPAPQPEQPAPAPKPEQPAPAPKPEQPAPAPK 483
> spn:SP_0117 surface protein A
Length=744
Score = 33.5 bits (75), Expect = 8.8, Method: Compositional matrix adjust.
Identities = 32/93 (34%), Positives = 41/93 (44%), Gaps = 18/93 (19%)
Query 39 ADMYREKFNLSDELSAEEIFDHVSEQAKKLKTRLEEIEKARKESQAKKHEPVPDKSTE-- 96
AD+ +E NL L + D + KL T+ E+EK +KE A +E PD E
Sbjct 391 ADLEKEISNLEILLGGADSEDDTAALQNKLATKKAELEKTQKELDAALNELGPDGDEEET 450
Query 97 --------------KP-QPQPQPQP-QPQPEPK 113
KP QP P P+P QP P PK
Sbjct 451 PAPAPQPEQPAPAPKPEQPAPAPKPEQPAPAPK 483
> spnn:T308_00570 choline binding protein J
Length=724
Score = 33.5 bits (75), Expect = 9.2, Method: Compositional matrix adjust.
Identities = 32/93 (34%), Positives = 41/93 (44%), Gaps = 18/93 (19%)
Query 39 ADMYREKFNLSDELSAEEIFDHVSEQAKKLKTRLEEIEKARKESQAKKHEPVPDKSTE-- 96
AD+ +E NL L + D + KL T+ E+EK +KE A +E PD E
Sbjct 380 ADLEKEISNLEILLGGADSEDDTAALQNKLATKKAELEKTQKELDAALNELGPDGDEEET 439
Query 97 --------------KP-QPQPQPQP-QPQPEPK 113
KP QP P P+P QP P PK
Sbjct 440 PAPAPQPEQPAPAPKPEQPAPAPKPEQPAPAPK 472
> snd:MYY_0199 surface protein A
Length=724
Score = 33.5 bits (75), Expect = 9.2, Method: Compositional matrix adjust.
Identities = 32/93 (34%), Positives = 41/93 (44%), Gaps = 18/93 (19%)
Query 39 ADMYREKFNLSDELSAEEIFDHVSEQAKKLKTRLEEIEKARKESQAKKHEPVPDKSTE-- 96
AD+ +E NL L + D + KL T+ E+EK +KE A +E PD E
Sbjct 380 ADLEKEISNLEILLGGADSEDDTAALQNKLATKKAELEKTQKELDAALNELGPDGDEEET 439
Query 97 --------------KP-QPQPQPQP-QPQPEPK 113
KP QP P P+P QP P PK
Sbjct 440 PAPAPQPEQPAPAPKPEQPAPAPKPEQPAPAPK 472
> snc:HMPREF0837_10423 psaA; manganese ABC transporter substrate-binding
lipoprotein
Length=724
Score = 33.5 bits (75), Expect = 9.2, Method: Compositional matrix adjust.
Identities = 32/93 (34%), Positives = 41/93 (44%), Gaps = 18/93 (19%)
Query 39 ADMYREKFNLSDELSAEEIFDHVSEQAKKLKTRLEEIEKARKESQAKKHEPVPDKSTE-- 96
AD+ +E NL L + D + KL T+ E+EK +KE A +E PD E
Sbjct 380 ADLEKEISNLEILLGGADSEDDTAALQNKLATKKAELEKTQKELDAALNELGPDGDEEET 439
Query 97 --------------KP-QPQPQPQP-QPQPEPK 113
KP QP P P+P QP P PK
Sbjct 440 PAPAPQPEQPAPAPKPEQPAPAPKPEQPAPAPK 472
> snt:SPT_0163 pneumococcal surface protein A
Length=724
Score = 33.5 bits (75), Expect = 9.2, Method: Compositional matrix adjust.
Identities = 32/93 (34%), Positives = 41/93 (44%), Gaps = 18/93 (19%)
Query 39 ADMYREKFNLSDELSAEEIFDHVSEQAKKLKTRLEEIEKARKESQAKKHEPVPDKSTE-- 96
AD+ +E NL L + D + KL T+ E+EK +KE A +E PD E
Sbjct 380 ADLEKEISNLEILLGGADSEDDTAALQNKLATKKAELEKTQKELDAALNELGPDGDEEET 439
Query 97 --------------KP-QPQPQPQP-QPQPEPK 113
KP QP P P+P QP P PK
Sbjct 440 PAPAPQPEQPAPAPKPEQPAPAPKPEQPAPAPK 472
Lambda K H a alpha
0.306 0.128 0.354 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 128140709664