bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: all_orgs
14,240,465 sequences; 5,121,972,263 total letters
Query= Contig-2_CDS_annotation_glimmer3.pl_2_4
Length=636
Score E
Sequences producing significant alignments: (Bits) Value
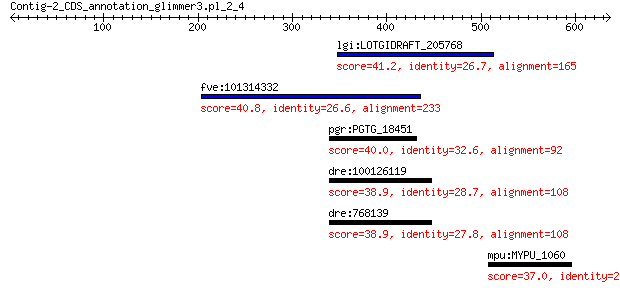
lgi:LOTGIDRAFT_205768 hypothetical protein 41.2 0.59
fve:101314332 capsid protein VP1-like 40.8 0.95
pgr:PGTG_18451 GDP-L-fucose synthase 40.0 1.3
dre:100126119 zgc:173683 38.9 2.9
dre:768139 zgc:153776 38.9 3.2
mpu:MYPU_1060 hemK; protoporphirogen oxidase HemK 37.0 7.8
> lgi:LOTGIDRAFT_205768 hypothetical protein
Length=318
Score = 41.2 bits (95), Expect = 0.59, Method: Compositional matrix adjust.
Identities = 44/187 (24%), Positives = 71/187 (38%), Gaps = 36/187 (19%)
Query 348 IRVAGGSGIPSSVAGSALGVRSLLGGEFSILALRQAEALQKWKEIT-QSVDTNYRDQIKA 406
I V GGSG L+G + + +KW ++ + D RD KA
Sbjct 8 ILVTGGSG--------------LVGKAIERVVQDEKRDDEKWIFVSSKDADLTDRDATKA 53
Query 407 HFGINTPASMSHMAQYIGGIARNLD-------ISEVVNNNL----SETGSEAVIYGKGVG 455
F + P + H+A ++GG+ RNL ++ ++N+N+ ETG + V+
Sbjct 54 MFAKHKPTHVIHLAAFVGGLFRNLKYNLDFFRVNSIINDNVLHTSYETGVKKVVSCLSTC 113
Query 456 TGSGKMRY-------HTGSQYCIIMCIYHAVPLLDYAISGQDSQLLCTSVEDLPIPEF-- 506
K Y H G + A ++D G +Q C +P F
Sbjct 114 IFPDKTTYPIDETMVHNGPPHSSNFGYSFAKRMIDVQNRGYHTQHGCNFTSVIPTNVFGP 173
Query 507 -DNIGME 512
DN +E
Sbjct 174 YDNFNLE 180
> fve:101314332 capsid protein VP1-like
Length=421
Score = 40.8 bits (94), Expect = 0.95, Method: Compositional matrix adjust.
Identities = 62/244 (25%), Positives = 92/244 (38%), Gaps = 58/244 (24%)
Query 203 SQNVSVSLFPLLAYQKIYQDFFRWSQWENADPTAYNVDYYNGSGNLFGNGGIASSIPSSN 262
S VS S P+ AY IY +FR +N+ VD +G + PS+N
Sbjct 170 SNTVSHSALPVRAYNLIYNQWFRDENLQNS----VVVDKGDG----------PDTTPSTN 215
Query 263 DYWKRDNMFSLRYCNWNKDMFMGLLPNSQFGDVAV-----------VSGVEGIDTFVPVE 311
++L D F LP Q G AV SG G D V
Sbjct 216 --------YTLLRRGKRHDYFTSALPWPQKGGTAVSLPLGTSAPIAFSGASGSD----VG 263
Query 312 VFNSINETNIAKPPLTGTHTPVYTDDAMTSSTTPSRIRVAGGSGIPSSVAGSALGVRSLL 371
V ++ I TG+ T + A VA G S A +A
Sbjct 264 VISTTQGNLIKNMYSTGSGTSLKIGSAT----------VATGLYADLSAATAA------- 306
Query 372 GGEFSILALRQAEALQKWKEITQSVDTNYRDQIKAHFGINTPASMSHMAQYIGGIARNLD 431
+I LRQ+ +QK E T Y + I++HFG+ +P + +Y+GG + ++
Sbjct 307 ----TINQLRQSFQIQKLLERDARGGTRYTEIIRSHFGVASPDARLQRPEYLGGGSTPIN 362
Query 432 ISEV 435
I+ +
Sbjct 363 IAPI 366
> pgr:PGTG_18451 GDP-L-fucose synthase
Length=337
Score = 40.0 bits (92), Expect = 1.3, Method: Compositional matrix adjust.
Identities = 30/94 (32%), Positives = 45/94 (48%), Gaps = 8/94 (9%)
Query 339 MTSSTTPSRIRVAGGSGIPSSVAGSALGVRSLLGGEFSILALRQAEALQKWKEITQSVDT 398
MTSS++P+ I V GGSG+ S + +G F R E +KW ++ S D
Sbjct 1 MTSSSSPAVILVTGGSGLVGSALKHVIDTEP-IGSRF---GRRTKE--EKWVFLSGSKDG 54
Query 399 NYR--DQIKAHFGINTPASMSHMAQYIGGIARNL 430
+ R DQ F P + H+A +GG+ N+
Sbjct 55 DLRELDQTLKVFEKYKPTHVIHLAALVGGLFANM 88
> dre:100126119 zgc:173683
Length=320
Score = 38.9 bits (89), Expect = 2.9, Method: Compositional matrix adjust.
Identities = 31/115 (27%), Positives = 53/115 (46%), Gaps = 20/115 (17%)
Query 339 MTSSTTPSRIRVAGGSGIPSSVAGSALGVRSLLGGEFSILALRQAEALQKWKEITQSVDT 398
M + P R+ V GGSG+ + G E++ L+ ++A L KE T+++
Sbjct 1 MNGTVEPMRVLVTGGSGLVGRAIERVVKEEGREGEEWTFLSSKEANLLSA-KE-TRAIFE 58
Query 399 NYRDQIKAHFGINTPASMSHMAQYIGGIAR----NLDI---SEVVNNNLSETGSE 446
YR P + H+A +GG+ R NLD + +N+N+ +T +E
Sbjct 59 KYR-----------PTHVIHLAAMVGGLFRNMRQNLDFWRNNVFINDNVLQTANE 102
> dre:768139 zgc:153776
Length=320
Score = 38.9 bits (89), Expect = 3.2, Method: Compositional matrix adjust.
Identities = 30/115 (26%), Positives = 51/115 (44%), Gaps = 20/115 (17%)
Query 339 MTSSTTPSRIRVAGGSGIPSSVAGSALGVRSLLGGEFSILALRQAEALQKWKEITQSVDT 398
M + P R+ V GGSG+ + G E++ L+ + A L E T+++
Sbjct 1 MNGTVEPMRVLVTGGSGLVGRAIERVVKDEGREGEEWTFLSSKDANLLSA--EETRAIFQ 58
Query 399 NYRDQIKAHFGINTPASMSHMAQYIGGIAR----NLDI---SEVVNNNLSETGSE 446
YR P + H+A +GG+ R NLD + +N+N+ +T +E
Sbjct 59 KYR-----------PTHVIHLAAMVGGLFRNMRQNLDFWRNNVFINDNVLQTANE 102
> mpu:MYPU_1060 hemK; protoporphirogen oxidase HemK
Length=230
Score = 37.0 bits (84), Expect = 7.8, Method: Compositional matrix adjust.
Identities = 25/89 (28%), Positives = 41/89 (46%), Gaps = 3/89 (3%)
Query 508 NIGMEAVPAITLFNSNAFDNDLESDFDFLGYNPRYWPWKSKIDRVHGAFLTTLKDWVAPI 567
N+ + V + S+ F+N ++ DFD + NP Y ++ KID+ F LK AP
Sbjct 116 NVKINNVKNYKIIQSDLFEN-IQGDFDIIVSNPPYLSYEQKIDK-SVKFFEPLKALYAPK 173
Query 568 DDFYL-NRWFASGGSSQASISWPFFKVNP 595
+ +Y + S FF++NP
Sbjct 174 NGWYFYEKIIEKASSFLKKDGMLFFEINP 202
Lambda K H a alpha
0.319 0.135 0.419 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 1491003542770