bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: all_orgs
14,240,465 sequences; 5,121,972,263 total letters
Query= Contig-30_CDS_annotation_glimmer3.pl_2_2
Length=555
Score E
Sequences producing significant alignments: (Bits) Value
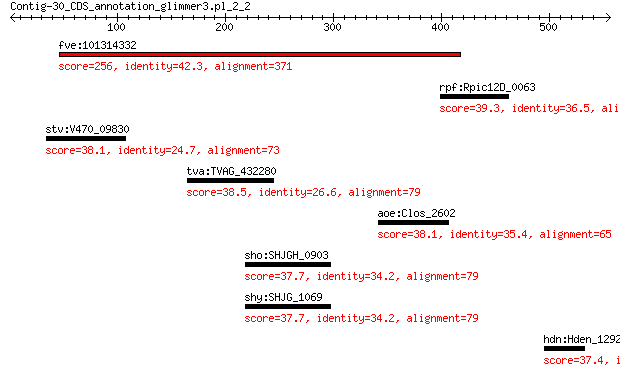
fve:101314332 capsid protein VP1-like 256 1e-75
rpf:Rpic12D_0063 hypothetical protein 39.3 2.4
stv:V470_09830 capsid protein 38.1 4.8
tva:TVAG_432280 Clan CA, family C19, ubiquitin hydrolase-like ... 38.5 4.9
aoe:Clos_2602 S-layer domain-containing protein 38.1 5.9
sho:SHJGH_0903 hypothetical protein 37.7 7.4
shy:SHJG_1069 hypothetical protein 37.7 7.4
hdn:Hden_1292 3-oxoacyl-(acyl-carrier-protein) synthase III (E... 37.4 7.8
> fve:101314332 capsid protein VP1-like
Length=421
Score = 256 bits (653), Expect = 1e-75, Method: Compositional matrix adjust.
Identities = 157/383 (41%), Positives = 220/383 (57%), Gaps = 50/383 (13%)
Query 46 VLPGDTFDIDTSKVIRMPSLLTPIMDNLYLDTYYFFVPNRIVWQHWKELMGENNESAWIP 105
VLPGDTF+++ + R+ + + P+MDNL+LD+++FFVPNR+VW +W + MGE + A
Sbjct 77 VLPGDTFNVNVTMFGRLATPIFPVMDNLHLDSFFFFVPNRLVWNNWVKFMGEQDNPA--D 134
Query 106 TTEYEVPQITAPSGGWSIGTIADYMGVPT----GVSG-LSVNALPFRAYALICNEWFRDE 160
+ Y +PQ +P+GG+++G++ DY G+PT GVS +S +ALP RAY LI N+WFRDE
Sbjct 135 SISYSIPQQVSPAGGYAVGSLQDYFGLPTAGQVGVSNTVSHSALPVRAYNLIYNQWFRDE 194
Query 161 NLCDPLNIPLTDATVAGVNTGTFVTDVAKGGLPYKAAKYRDYFTSCLPAPQKS-EDVTIP 219
NL + + + D T T + +G K DYFTS LP PQK V++P
Sbjct 195 NLQNSVVVDKGDG--PDTTPSTNYTLLRRG-------KRHDYFTSALPWPQKGGTAVSLP 245
Query 220 VSSGANYPVLSLSDIVPTPGTVPVKWNDANN----VVSDAQWLLGGKNYNGTITSNDISL 275
+ + A P+ ++ A+ V+S Q G + N S
Sbjct 246 LGTSA-----------------PIAFSGASGSDVGVISTTQ---------GNLIKNMYST 279
Query 276 TKTNTGPTYSAVTPINLWAVNDGSVSSATINQLRLAFQVQKLYERDARGGTRYIEVLKSH 335
+ SA L+A + + ++ATINQLR +FQ+QKL ERDARGGTRY E+++SH
Sbjct 280 GSGTSLKIGSATVATGLYA-DLSAATAATINQLRQSFQIQKLLERDARGGTRYTEIIRSH 338
Query 336 FGVTSPDARLQRPEYLGGNRIPIVISEI--NQTSGTSANSTPQGNPSGQSRTTDVHSDFK 393
FGV SPDARLQRPEYLGG PI I+ I +G +TPQGN + F
Sbjct 339 FGVASPDARLQRPEYLGGGSTPINIAPIAQTGGTGAQGTTTPQGNLAAFGTYMAKGHGFS 398
Query 394 KSFVEHGFIIGVMVARYDHTYQQ 416
+SFVEHG +IG++ R D TYQQ
Sbjct 399 QSFVEHGHVIGLVSVRADLTYQQ 421
> rpf:Rpic12D_0063 hypothetical protein
Length=862
Score = 39.3 bits (90), Expect = 2.4, Method: Compositional matrix adjust.
Identities = 23/63 (37%), Positives = 35/63 (56%), Gaps = 1/63 (2%)
Query 399 HGFIIGVMVARYDHTYQQGLERFWSRKGRLDYYWPVFANIGEQAVLNKEIYAQGNGTDDE 458
H F+ VM A D + + +R W+RKGRL + IG AV ++E +A+GN T
Sbjct 717 HPFVARVMRAMPDADWAERGQR-WARKGRLVGFLGALVGIGWDAVHSREEFAKGNQTLGY 775
Query 459 VFG 461
++G
Sbjct 776 LYG 778
> stv:V470_09830 capsid protein
Length=427
Score = 38.1 bits (87), Expect = 4.8, Method: Compositional matrix adjust.
Identities = 18/74 (24%), Positives = 39/74 (53%), Gaps = 1/74 (1%)
Query 34 NVGDIVPFYVDEVLPGDTFDIDTSKVIRMPSLLTPIMDNLYLDTYYFFVPNRIVW-QHWK 92
+G ++ V+ GD+F++D +R+ L + + +D + F+VP+R V+ + W
Sbjct 24 QIGRLITISTTPVIAGDSFEMDAVGALRLSPLRRGLAIDSTVDIFTFYVPHRHVYGEQWI 83
Query 93 ELMGENNESAWIPT 106
+ M + + +PT
Sbjct 84 KFMKDGVNATPLPT 97
> tva:TVAG_432280 Clan CA, family C19, ubiquitin hydrolase-like
cysteine peptidase
Length=1876
Score = 38.5 bits (88), Expect = 4.9, Method: Compositional matrix adjust.
Identities = 21/79 (27%), Positives = 40/79 (51%), Gaps = 2/79 (3%)
Query 165 PLNIPLTDATVAGVNTGTFVTDVAKGGLPYKAAKYRDYFTSCLPAPQKSEDVTIPVSSGA 224
P PLT ++ T + K + ++A KY +F +CL +P+KSE + +S
Sbjct 683 PFGCPLTSLHLSSTETLFVSIEKTKDKILHQA-KYEPHFLTCLSSPEKSEILANLLSQSH 741
Query 225 NYPVLSLSDIVPTPGTVPV 243
N + +++++ P +PV
Sbjct 742 NIQLFRVAELI-DPSNIPV 759
> aoe:Clos_2602 S-layer domain-containing protein
Length=493
Score = 38.1 bits (87), Expect = 5.9, Method: Compositional matrix adjust.
Identities = 23/67 (34%), Positives = 35/67 (52%), Gaps = 2/67 (3%)
Query 341 PDARLQRPEYLGG--NRIPIVISEINQTSGTSANSTPQGNPSGQSRTTDVHSDFKKSFVE 398
PD + R E+ I ++E+ +T G+S N TP+ +P TTD + D+ K+ V
Sbjct 319 PDVPMNREEFTRAVMRASAIRLAEVPKTRGSSRNKTPEISPFSDVATTDANYDYIKNGVA 378
Query 399 HGFIIGV 405
G I GV
Sbjct 379 KGIIQGV 385
> sho:SHJGH_0903 hypothetical protein
Length=512
Score = 37.7 bits (86), Expect = 7.4, Method: Compositional matrix adjust.
Identities = 27/79 (34%), Positives = 39/79 (49%), Gaps = 5/79 (6%)
Query 218 IPVSSGANYPVLSLSDIVPTPGTVPVKWNDANNVVSDAQWLLGGKNYNGTITSNDISLTK 277
+ VSSG++ L S +PGT + W + N S+ QW + N +GT+TS L
Sbjct 429 LTVSSGSSRLCLDASGQGTSPGTKVITW--SCNGQSNQQWNV---NADGTVTSAQSGLCL 483
Query 278 TNTGPTYSAVTPINLWAVN 296
TG + TP+ LW N
Sbjct 484 DVTGAATANGTPVQLWTCN 502
> shy:SHJG_1069 hypothetical protein
Length=512
Score = 37.7 bits (86), Expect = 7.4, Method: Compositional matrix adjust.
Identities = 27/79 (34%), Positives = 39/79 (49%), Gaps = 5/79 (6%)
Query 218 IPVSSGANYPVLSLSDIVPTPGTVPVKWNDANNVVSDAQWLLGGKNYNGTITSNDISLTK 277
+ VSSG++ L S +PGT + W + N S+ QW + N +GT+TS L
Sbjct 429 LTVSSGSSRLCLDASGQGTSPGTKVITW--SCNGQSNQQWNV---NADGTVTSAQSGLCL 483
Query 278 TNTGPTYSAVTPINLWAVN 296
TG + TP+ LW N
Sbjct 484 DVTGAATANGTPVQLWTCN 502
> hdn:Hden_1292 3-oxoacyl-(acyl-carrier-protein) synthase III
(EC:2.3.1.41)
Length=326
Score = 37.4 bits (85), Expect = 7.8, Method: Compositional matrix adjust.
Identities = 17/37 (46%), Positives = 24/37 (65%), Gaps = 0/37 (0%)
Query 495 DDYSKLPSLSDSWVQEDSAVVNRVIAVSEENSNQLWA 531
+D SK+ SD+W+QE S +VNR IA E ++ L A
Sbjct 24 EDLSKIVDTSDAWIQERSGIVNRHIADETEKTSDLGA 60
Lambda K H a alpha
0.316 0.133 0.410 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 1253412983580