bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: all_orgs
14,240,465 sequences; 5,121,972,263 total letters
Query= Contig-31_CDS_annotation_glimmer3.pl_2_5
Length=290
Score E
Sequences producing significant alignments: (Bits) Value
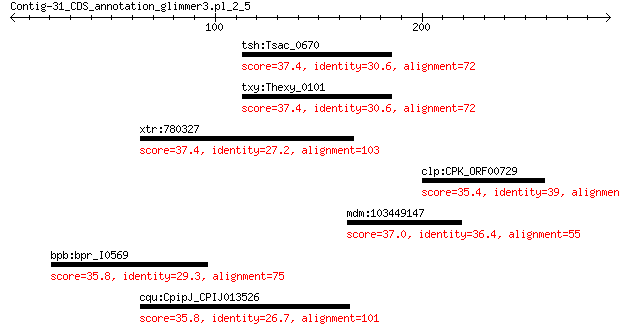
tsh:Tsac_0670 ParA/MinD-like ATPase 37.4 3.2
txy:Thexy_0101 ParA/MinD-like ATPase 37.4 3.4
xtr:780327 anapc1, apc1, mcpr, tsg24; anaphase promoting compl... 37.4 3.7
clp:CPK_ORF00729 hypothetical protein 35.4 3.9
mdm:103449147 ARF guanine-nucleotide exchange factor GNL2-like 37.0 5.6
bpb:bpr_I0569 ribonuclease Z 35.8 7.7
cqu:CpipJ_CPIJ013526 hypothetical protein 35.8 9.5
> tsh:Tsac_0670 ParA/MinD-like ATPase
Length=351
Score = 37.4 bits (85), Expect = 3.2, Method: Compositional matrix adjust.
Identities = 22/73 (30%), Positives = 34/73 (47%), Gaps = 1/73 (1%)
Query 113 IPNFVSAKVNRPYMLEHLHFIEAERYKALSLRYPNFGSKFRPYILRSILRKSPLQRF-KD 171
IP + RPY L+ + E+Y + NF + P I R L L++F D
Sbjct 147 IPRLLGIVGERPYALDENTILPIEKYGIKVISMGNFADEDTPLIWRGPLLGGVLEQFMND 206
Query 172 EYFEELVWMLPEL 184
Y+ EL +M+ +L
Sbjct 207 VYWGELDYMIIDL 219
> txy:Thexy_0101 ParA/MinD-like ATPase
Length=352
Score = 37.4 bits (85), Expect = 3.4, Method: Compositional matrix adjust.
Identities = 22/73 (30%), Positives = 34/73 (47%), Gaps = 1/73 (1%)
Query 113 IPNFVSAKVNRPYMLEHLHFIEAERYKALSLRYPNFGSKFRPYILRSILRKSPLQRF-KD 171
IP + RPY L+ + E+Y + NF + P I R L L++F D
Sbjct 147 IPRLLGIVGERPYALDENTILPIEKYGIKVISMGNFADEDTPLIWRGPLLGGVLEQFMND 206
Query 172 EYFEELVWMLPEL 184
Y+ EL +M+ +L
Sbjct 207 VYWGELDYMIIDL 219
> xtr:780327 anapc1, apc1, mcpr, tsg24; anaphase promoting complex
subunit 1
Length=1948
Score = 37.4 bits (85), Expect = 3.7, Method: Composition-based stats.
Identities = 28/103 (27%), Positives = 49/103 (48%), Gaps = 6/103 (6%)
Query 64 LTYNSEYVPKMSLTQIDDYLTEWLPVRPPKSIGTQLVARMVMDSRVNKKIPNFVSAKVNR 123
L NSE++P M T ID+ L +WL ++ + L A+ V DS+++ V V
Sbjct 1842 LFMNSEFLPIMKCT-IDNTLDQWLQANGDVALHSYLAAQAVEDSQLSMLACFLVYHSVPE 1900
Query 124 PYMLEHLHFIEAERYKALSLRYPNFGSKFRPYILRSILRKSPL 166
P L +E + L L++ + +R++LR +P+
Sbjct 1901 PKQLSEGGLQGSESFSELLLKFKDLKMP-----VRALLRLAPI 1938
> clp:CPK_ORF00729 hypothetical protein
Length=121
Score = 35.4 bits (80), Expect = 3.9, Method: Compositional matrix adjust.
Identities = 23/59 (39%), Positives = 29/59 (49%), Gaps = 9/59 (15%)
Query 200 FPQFKGLLKYINIRDYQLFSKRLRKYLSKKIGKYEKIHSYVVSEYSPKTLRPHFHILFF 258
PQ+ L+K QLF KRLR +S KI + EY K RPH+H+L F
Sbjct 64 LPQYGSLVKL----HLQLFLKRLRDRISP-----HKIRYFGCGEYGTKLQRPHYHLLIF 113
> mdm:103449147 ARF guanine-nucleotide exchange factor GNL2-like
Length=1384
Score = 37.0 bits (84), Expect = 5.6, Method: Compositional matrix adjust.
Identities = 20/55 (36%), Positives = 31/55 (56%), Gaps = 8/55 (15%)
Query 164 SPLQRFKDEYFEELVWMLPELAESLKKKNNTDANGAFPQFKGLLKYINIRDYQLF 218
SPL + + FE LV M+ +A+S+ K+N+T +G +P I IR+Y F
Sbjct 433 SPLTTLQIQAFEGLVIMIHNIADSIDKENDTSPSGPYP--------IEIREYTPF 479
> bpb:bpr_I0569 ribonuclease Z
Length=306
Score = 35.8 bits (81), Expect = 7.7, Method: Compositional matrix adjust.
Identities = 22/75 (29%), Positives = 35/75 (47%), Gaps = 6/75 (8%)
Query 21 KYTGDFVKVDCGECPYCLIKKSDRATQKCDFVKFNHKYCYFVTLTYNSEYVPKMSLTQID 80
+Y G V +DCGE +KK +++ D + F H + V+ +P M LT +
Sbjct 26 RYNGKCVLIDCGEATQIAMKKKGLSSKPIDVICFTHFHADHVS------GLPGMLLTMGN 79
Query 81 DYLTEWLPVRPPKSI 95
TE L + PK +
Sbjct 80 AERTEPLLIVGPKGV 94
> cqu:CpipJ_CPIJ013526 hypothetical protein
Length=451
Score = 35.8 bits (81), Expect = 9.5, Method: Compositional matrix adjust.
Identities = 27/101 (27%), Positives = 44/101 (44%), Gaps = 12/101 (12%)
Query 64 LTYNSEYVPKMSLTQIDDYLTEWLPVRPPKSIGTQLVARMVMDSRVNKKIPNFVSAKVNR 123
+ N EY+P+M +T + D I Q ++ V D V ++ + SA V +
Sbjct 26 MDLNKEYLPEMGITTMRDI------------IAIQRHSKTVCDQSVRDRVLSSQSAVVEK 73
Query 124 PYMLEHLHFIEAERYKALSLRYPNFGSKFRPYILRSILRKS 164
P + EHL + + +L P S RP +S+ KS
Sbjct 74 PELPEHLTNLVPVAAVSATLSNPTKSSVSRPSSAKSLPSKS 114
Lambda K H a alpha
0.324 0.139 0.433 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 479831790136